Notre recherche
Notre équipe utilise des méthodes de bioinformatique et de biologie structurale afin de comprendre les principes qui régissent les structures protéiques, les interactions biomoléculaires et les mécanismes moléculaires des processus cellulaires. Actuellement, la bioinformatique joue un rôle d’importance croissante dans la recherche en biologie. La grande quantité de données issues du séquençage obtenues ces dernières années pose de nouveaux défis pour les bioinformaticiens: déchiffrer le rôle biologique de millions de séquences protéiques.Pour relever ce défi, notre équipe a pour objectif le développement de nouvelles méthodes informatiques permettant la mise en place de la chaîne de traitement nécessaire pour l’annotation structurale et fonctionnelle des protéomes.
Les thèmes principaux de notre recherche sont :
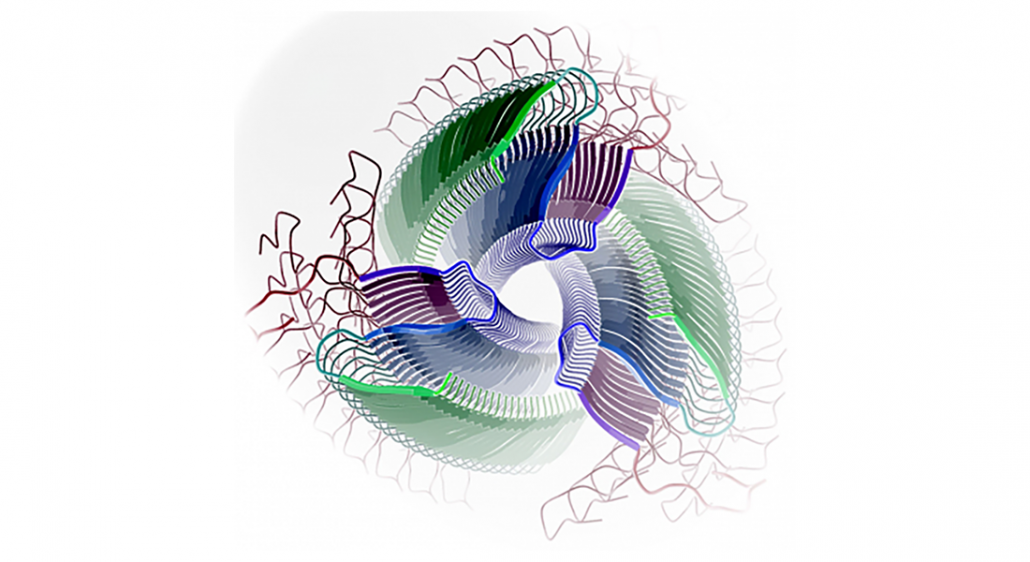
Protéines à unités répétitives en tandem
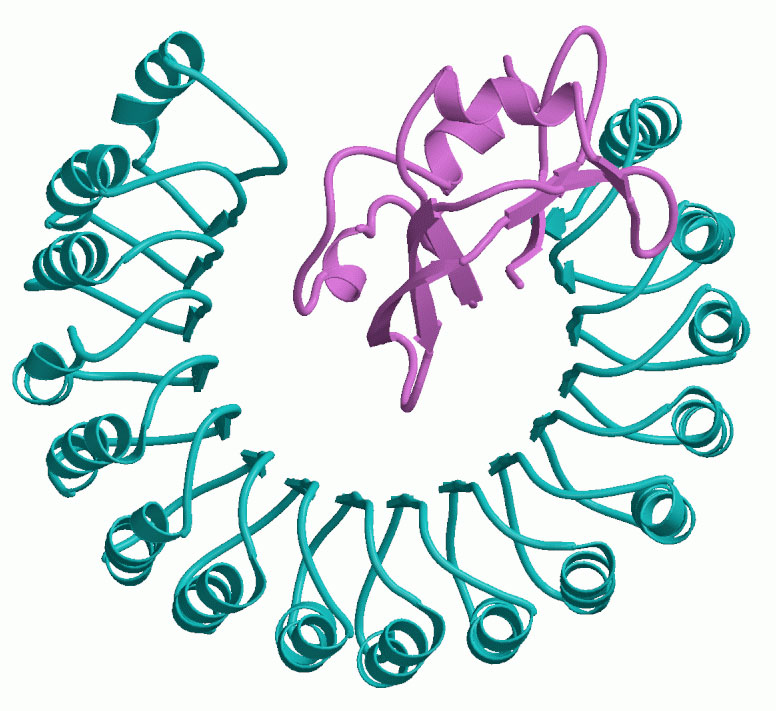
Nous avons initié l’analyse bioinformatique, la classification, la prédiction structurale et la modélisation de protéines à séquences répétitives. Le développement ultérieur de méthodes fiables d’identification des motifs répétitifs protéiques et la prédiction ab initio de structure 3D promet d’être un sujet de recherche fertile en bioinformatique structurale. Au cours des dernières années, de nombreuses preuves ont été accumulées sur la forte présence des répétitions en tandem dans les séquences en acides aminés des facteurs de virulence de pathogènes et de certains amyloïdes et prions. La découverte et les prédictions structure-fonction de ces domaines peuvent conduire à l’identification de cibles pour de nouveaux antibiotiques et vaccins contre des maladies infectieuses émergeantes et au développement d’inhibiteurs de l’amyloïdogénèse.
Amyloides et Prions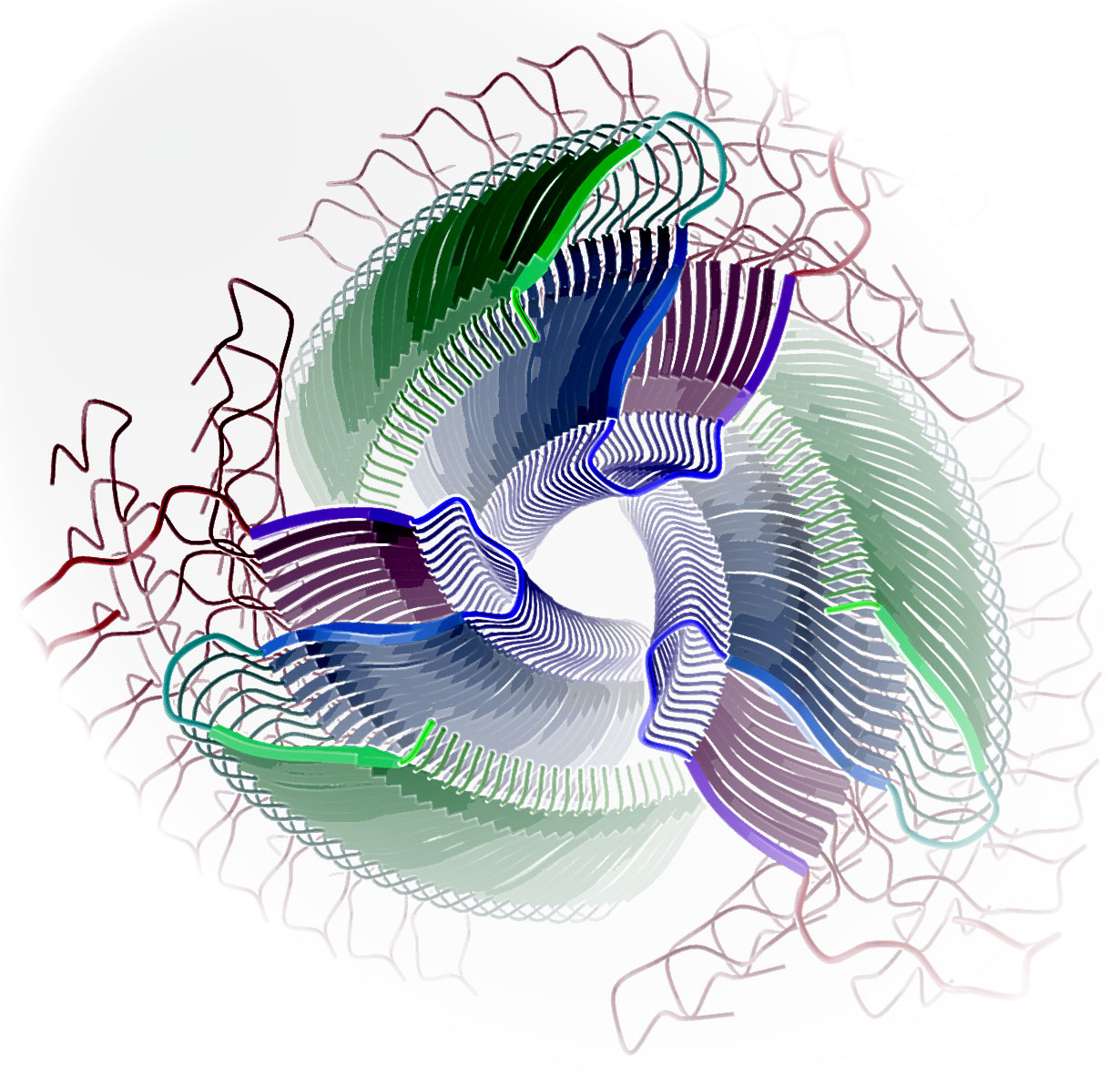
De nombreux travaux ont suggéré que les fibrilles amyloïdes jouent un rôle causal dans certaines pathologies neurodégénératives, y compris les maladies d’Alzheimer, de Parkinson et de Hungtington. La détermination de la structure des fibrilles se fait au moyen de suggestions de modèles structuraux basés sur l’accumulation de résultats expérimentaux, jusqu’à ce qu’un modèle unique permette d’expliquer l’ensemble des données. Nous contribuons aux avancées dans ce domaine en développant des modèles structuraux de fibrilles amyloïdes et de prions. Notre but ultérieur sera la prédiction des propriétés amyloïdogéniques d’une séquence en acides aminés à partir de sa structure ainsi que le design d’inhibiteurs de la fibrillogénèse.
Pour accéder à nos outils bioinformatiques aller sur : http://bioinfo.montp.cnrs.fr/
En savoir plus
Financements


Publications
2025
- Developing machine-learning-based amyloidogenicity predictors with Cross-Beta DB. Gonay V, Dunne MP, Caceres-Delpiano J, Kajava AV. Alzheimers Dement. 2025 Jan 8. Pubmed
- RepeatsDB in 2025: expanding annotations of structured tandem repeats proteins on AlphaFoldDB. Clementel D, Arrías PN, Mozaffari S, Osmanli Z, Castro XA; RepeatsDB curators; Ferrari C, Kajava AV, Tosatto SCE, Monzon AM. Nucleic Acids Res. 2025 Jan 6;53(D1):D575-D581. Pubmed
2024
- Cross-reactivity of rPvs48/45, a recombinant Plasmodium vivax protein, with sera from Plasmodium falciparum endemic areas of Africa. Balam S, Miura K, Ayadi I, Konaté D, Incandela NC, Agnolon V, Guindo MA, Diakité SAS, Olugbile S, Nebie I, Herrera SM, Long C, Kajava AV, Diakité M, Corradin G, Herrera S, Herrera MA. bioRxiv [Preprint]. 2024 Apr 15:2024.04.10.588966. Pubmed
- AmyloComp: A Bioinformatic Tool for Prediction of Amyloid Co-aggregation. Bondarev SA, Uspenskaya MV, Leclercq J, Falgarone T, Zhouravleva GA, Kajava AV. J Mol Biol. 2024 Jan 5:168437. Pubmed
- Quantitative model and physical mechanisms of iRBC membrane curling during egress of malaria parasites. N. Gorkavyi, A. Parmeggiani, A. V. Kajava (2024) bioRxiv [Preprint]. 2024 doi: https://doi.org/10.1101/2024.02.13.579773
- Frameshifts in Tandem Repeats: Consequences on Protein Physicochemical Properties and Function Z Osmanli, G Aldrian, J Leclercq, T Falgarone, SM. Gomez Bergna, DN. Prada Gori, AV Oleinikov, I Shahmuradov, AV Kajava. bioRxiv. 2024 doi: https://doi.org/10.1101/2024.06.02.597034
2023
- A STRP-ed definition of Structured Tandem Repeats in Proteins. Monzon AM, Arrías PN, Elofsson A, Mier P, Andrade-Navarro MA, Bevilacqua M, Clementel D, Bateman A, Hirsh L, Fornasari MS, Parisi G, Piovesan D, Kajava AV, Tosatto SCE. J Struct Biol. 2023 Aug 29;215(4):108023. Pubmed
- The repetitive structure of DNA clamps: An overlooked protein tandem repeat. Arrías PN, Monzon AM, Clementel D, Mozaffari S, Piovesan D, Kajava AV, Tosatto SCE. J Struct Biol. 2023 Sep;215(3):108001.. Pubmed
- Why does the first protein repeat often become the only one? Manasra S, Kajava AV. J Struct Biol. 2023 Sep;215(3):108014. Pubmed
- Census of exposed aggregation-prone regions in proteomes. Falgarone T, Villain E, Richard F, Osmanli Z, Kajava AV. Brief Bioinform. 2023 Jul 20;24(4):bbad183. Pubmed
- ATR-FTIR spectrum analysis of plasma samples for rapid identification of recovered COVID-19 individuals. Karas BY, Sitnikova VE, Nosenko TN, Dedkov VG, Arsentieva NA, Gavrilenko NV, Moiseev IS, Totolian AA, Kajava AV, Uspenskaya MV. J Biophotonics. 2023 Jul;16(7):e202200166. Pubmed
- Pathogenic TRIO variants associated with neurodevelopmental disorders perturb the molecular regulation of TRIO and axon pathfinding in vivo. Bonnet M, Roche F, Fagotto-Kaufmann C, Gazdagh G, Truong I, Comunale F, Barbosa S, Bonhomme M, Nafati N, Hunt D, Rodriguez MP, Chaudhry A, Shears D, Madruga M, Vansenne F, Curie A, Kajava AV, Baralle D, Fassier C, Debant A, Schmidt S. Mol Psychiatry. 2023 Apr;28(4):1527-1544. Pubmed
2022
- Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies. Nilsson JF, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava AV, Longhi S. Int J Mol Sci. 2022 Dec 26;24(1):399. Pubmed
- The Difference in Structural States between Canonical Proteins and Their Isoforms Established by Proteome-Wide Bioinformatics Analysis. Osmanli Z, Falgarone T, Samadova T, Aldrian G, Leclercq J, Shahmuradov I, Kajava AV. Biomolecules. 12(11):1610. Pubmed
- A structural biology community assessment of AlphaFold2 applications. Akdel M, Pires DEV, Pardo EP, Jänes J, Zalevsky AO, Mészáros B, Bryant P, Good LL, Laskowski RA, Pozzati G, Shenoy A, Zhu W, Kundrotas P, Serra VR, Rodrigues CHM, Dunham AS, Burke D, Borkakoti N, Velankar S, Frost A, Basquin J, Lindorff-Larsen K, Bateman A, Kajava AV, Valencia A, Ovchinnikov S, Durairaj J, Ascher DB, Thornton JM, Davey NE, Stein A, Elofsson A, Croll TI, Beltrao P. Nat Struct Mol Biol. 29(11):1056-1067. Pubmed
- A SARS-CoV-2 Spike Receptor Binding Motif Peptide Induces Anti-Spike Antibodies in Mice and Is Recognized by COVID-19 Patients. Pratesi F, Errante F, Pacini L, Peña-Moreno IC, Quiceno S, Carotenuto A, Balam S, Konaté D, Diakité MM, Arévalo-Herrera M, Kajava AV, Rovero P, Corradin G, Migliorini P, Papini AM, Herrera S. Front Immunol. 13:879946. Pubmed
- Seroreactivity of the Severe Acute Respiratory Syndrome Coronavirus 2 Recombinant S Protein, Receptor-Binding Domain, and Its Receptor-Binding Motif in COVID-19 Patients and Their Cross-Reactivity With Pre-COVID-19 Samples From Malaria-Endemic Areas. Traoré A, Guindo MA, Konaté D, Traoré B, Diakité SA, Kanté S, Dembélé A, Cissé A, Incandela NC, Kodio M, Coulibaly YI, Faye O, Kajava AV, Pratesi F, Migliorini P, Papini AM, Pacini L, Rovero P, Errante F, Diakité M, Arevalo-Herrera M, Herrera S, Corradin G, Balam S. Front Immunol. 2022 Apr 27;13:856033. Pubmed
- Predicting Protein Conformational Disorder and Disordered Binding Sites. Tamburrini KC, Pesce G, Nilsson J, Gondelaud F, Kajava AV, Berrin JG, Longhi S. Methods Mol Biol. 2449:95-147. Pubmed
- TAPASS: Tool for annotation of protein amyloidogenicity in the context of other structural states. Falgarone T, Villain É, Guettaf A, Leclercq J, Kajava AV. J Struct Biol. 214(1):107840. Pubmed
- The C-terminal segment of Leishmania major HslU: Toward potential inhibitors of LmHslVU activity. Singh P, Samanta K, Kebe NM, Michel G, Legrand B, Sitnikova VE, Kajava AV, Pagès M, Bastien P, Pomares C, Coux O, Hernandez JF. Bioorg Chem. 119:105539. Pubmed
- Aspartate-phobia of thermophiles as a reaction to deleterious chemical transformations. Villain E, Fort P, Kajava AV. Bioessays. 44(1):e2100213. Pubmed
2021
- Identification of a Region in the Common Amino-terminal Domain of Hendra Virus P, V, and W Proteins Responsible for Phase Transition and Amyloid Formation. Salladini E, Gondelaud F, Nilsson JF, Pesce G, Bignon C, Murrali MG, Fabre R, Pierattelli R, Kajava AV, Horvat B, Gerlier D, Mathieu C, Longhi S. Biomolecules 1, 1324 Pubmed
- Critical assessment of protein intrinsic disorder prediction. Necci M, Piovesan D; CAID Predictors; DisProt Curators, Tosatto SCE. Nat Methods. 18(5):472-481. Pubmed
- RepeatsDB in 2021: improved data and extended classification for protein tandem repeat structures. Paladin L, Bevilacqua M, Errigo S, Piovesan D, Mičetić I, Necci M, Monzon AM, Fabre ML, Lopez JL, Nilsson JF, Rios J, Menna PL, Cabrera M, Buitron MG, Kulik MG, Fernandez-Alberti S, Fornasari MS, Parisi G, Lagares A, Hirsh L, Andrade-Navarro MA, Kajava AV, Tosatto SCE. Nucleic Acids Res. 49(D1):D452-D457 Pubmed
- Immunoreactivity of Sera From Low to Moderate Malaria-Endemic Areas Against Plasmodium vivax rPvs48/45 Proteins Produced in Escherichia coli and Chinese Hamster Ovary Systems. Arévalo-Herrera M, Miura K, Cespedes N, Echeverry C, Solano E, Castellanos A, Ramirez JS, Miranda A, Kajava AV, Long C, Corradin G, Herrera S. Front Immunol. 2021 Jun 24;12:634738. Pubmed
- Δ133p53β isoform pro-invasive activity is regulated through an aggregation-dependent mechanism in cancer cells. Arsic N, Slatter T, Gadea G, Villain E, Fournet A, Kazantseva M, Allemand F, Sibille N, Seveno M, de Rossi S, Mehta S, Urbach S, Bourdon JC, Bernado P, Kajava AV, Braithwaite A, Roux P. Nat Commun. 2021 Sep 15;12(1):5463. Pubmed
2020
- Estimation of amyloid aggregate sizes with semi-denaturing detergent agarose gel electrophoresis and its limitations. Drozdova PB, Barbitoff YA, Belousov MV, Skitchenko RK, Rogoza TM, Leclercq JY, Kajava AV, Matveenko AG, Zhouravleva GA, Bondarev SA Prion. 14(1):118-128. Pubmed
- Trimeric SARS-CoV-2 Spike Proteins Produced from CHO Cells in Bioreactors Are High-Quality Antigens. P. Pino, J. Kint, D. Kiseljak, V. Agnolon, G. Corradin, A. V. Kajava, P. Rovero, R. Dijkman, G. den Hartog,[…] Processes, 8(12), 1539 Pubmed
- Multifunctional Amyloid Oligomeric Nanoparticles for Specific Cell Targeting and Drug Delivery. Wang W, Azizyan RA, Garro A, Kajava AV, Ventura S. Biomacromolecules. 21(10):4302-4312. Pubmed
- Modeling polymorphic ventricular tachycardia at rest using patient-specific induced pluripotent stem cell-derived cardiomyocytes. Sleiman Y, Souidi M, Kumar R, Yang E, Jaffré F, Zhou T, Bernardin A, Reiken S, Cazorla O, Kajava AV, Moreau A, Pasquié JL, Marks AR, Lerman BB, Chen S, Cheung JW, Evans T, Lacampagne A, Meli AC. EBioMedicine. 2020 Oct;60:103024. Pubmed
- Amyloidogenicity as a driving force for the formation of functional oligomers. Azizyan RA, Wang W, Anikeenko A, Radkova Z, Bakulina A, Garro A, Charlier L, Dumas C, Ventura S, Kajava AV. J Struct Biol. 212(1):107604. Pubmed
- Point mutations affecting yeast prion propagation change the structure of its amyloid fibrils. A. I. Sulatskayaa, S. A. Bondarev, M. I. Sulatsky, N. P. Trubitsina, M. V.Belousov, G. A. Zhouravleva, M. A. Llanos,[…] Journal of Molecular Liquids. 314, 113618. Pubmed
- Tally-2.0: upgraded validator of tandem repeat detection in protein sequences. Perovic V, Leclercq JY, Sumonja N, Richard FD, Veljkovic N, Kajava AV. Bioinformatics. 36(10):3260-3262. Pubmed
- Census of halide-binding sites in protein structures. Skitchenko RK, Usoltsev D, Uspenskaya M, Kajava AV, Guskov A. Bioinformatics. 36(10):3064-3071. Pubmed
- FTIR Spectroscopy Study of the Secondary Structure Changes in Human Serum Albumin and Trypsin under Neutral Salts. Usoltsev D, Sitnikova V, Kajava A, Uspenskaya M. Biomolecules. 10(4):606. Pubmed
- Opposite Modulation of RAC1 by Mutations in TRIO Is Associated with Distinct, Domain-Specific Neurodevelopmental Disorders. Barbosa S, Greville-Heygate S, Bonnet M, Godwin A, Fagotto-Kaufmann C, Kajava AV, Laouteouet D, Mawby R, Wai HA, Dingemans AJM, Hehir-Kwa J, Willems M, Capri Y, Mehta SG, Cox H, Goudie D, Vansenne F, Turnpenny P, Vincent M, Cogné B, Lesca G, Hertecant J, Rodriguez D, Keren B, Burglen L, Gérard M, Putoux A; C4RCD Research Group, Cantagrel V, Siquier-Pernet K, Rio M, Banka S, Sarkar A, Steeves M, Parker M, Clement E, Moutton S, Tran Mau-Them F, Piton A, de Vries BBA, Guille M, Debant A, Schmidt S, Baralle D. Am J Hum Genet. pii: S0002-9297(20)30018-5. Pubmed
- DisProt: intrinsic protein disorder annotation in 2020. Hatos A, Hajdu-Soltész B, Monzon AM, Palopoli N, Álvarez L, Aykac-Fas B, Bassot C, Benítez GI, Bevilacqua M, Chasapi A,[…] Nucleic Acids Res. pii: gkz975. Pubmed
- Disentangling the complexity of low complexity proteins. Mier P, Paladin L, Tamana S, Petrosian S, Hajdu-Soltész B, Urbanek A, Gruca A, Plewczynski D, Grynberg M, Bernadó P, Gáspári Z, Ouzounis CA, Promponas VJ, Kajava AV, Hancock JM, Tosatto SCE, Dosztanyi Z, Andrade-Navarro MA. Brief Bioinform. 2020 Mar 23;21(2):458-472. Pubmed
2019
- Design of a New [PSI +]-No-More Mutation in SUP35 With Strong Inhibitory Effect on the [PSI +] Prion Propagation. Danilov LG, Matveenko AG, Ryzhkova VE, Belousov MV, Poleshchuk OI, Likholetova DV, Sokolov PA, Kasyanenko NA, Kajava AV, Zhouravleva GA, Bondarev SA. Front Mol Neurosci. 12:274. Pubmed
- Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Tørresen OK, Star B, Mier P, Andrade-Navarro MA, Bateman A, Jarnot P, Gruca A, Grynberg M, Kajava AV, Promponas VJ, Anisimova M, Jakobsen KS, Linke D. Nucleic Acids Res. 47(21):10994-11006 Pubmed
- Prion soft amyloid core driven self-assembly of globular proteins into bioactive nanofibrils. Wang W, Navarro S, Azizyan RA, Baño-Polo M, Esperante SA, Kajava AV, Ventura S. Nanoscale. 11(26):12680-12694. Pubmed
- Porins and Amyloids are Coded by Similar Sequence Motifs. Villain E, Nikekhin AA, Kajava AV. Proteomics. 2019 Mar;19(6):e1800075. Pubmed
- The HslV Protease from Leishmania major and Its Activation by C-terminal HslU Peptides. Kebe NM, Samanta K, Singh P, Lai-Kee-Him J, Apicella V, Payrot N, Lauraire N, Legrand B, Lisowski V, Mbang-Benet DE, Pages M, Bastien P, Kajava AV, Bron P, Hernandez JF, Coux O. Int J Mol Sci. 20(5). Pubmed
- Systematic FTIR Spectroscopy Study of the Secondary Structure Changes in Human Serum Albumin under Various Denaturation Conditions. Usoltsev D, Sitnikova V, Kajava A and Uspenskaya M. Biomolecules 2019, 9(8). pii: E359. Pubmed
2018
- Post-Translational Modifications and Diastolic Calcium Leak Associated to the Novel RyR2-D3638A Mutation Lead to CPVT in Patient-Specific hiPSC-Derived Cardiomyocytes. Acimovic I, Refaat MM, Moreau A, Salykin A, Reiken S, Sleiman Y, Souidi M, Přibyl J, Kajava AV, Richard S, Lu JT, Chevalier P, Skládal P, Dvořak P, Rotrekl V, Marks AR, Scheinman MM, Lacampagne A, Meli AC. J Clin Med. 8;7(11) Pubmed
- Protein Co-Aggregation Related to Amyloids: Methods of Investigation, Diversity, and Classification. Bondarev SA, Antonets KS, Kajava AV, Nizhnikov AA, Zhouravleva GA. Int J Mol Sci. ; 19(8) Pubmed
- BetaSerpentine: a bioinformatics tool for reconstruction of amyloid structures. Bondarev SA, Bondareva OV, Zhouravleva GA, Kajava AV. Bioinformatics. 34(4):599-608 Pubmed
- Editorial for special issue « Proteins with tandem repeats: sequences, structures and functions ». Kajava AV, Tosatto SCE. J Struct Biol. 201(2):86-87. Pubmed
- Identification of the Autochaperone Domain in the Type Va Secretion System (T5aSS): Prevalent Feature of Autotransporters with a β-Helical Passenger. Rojas-Lopez M, Zorgani MA, Kelley LA, Bailly X, Kajava AV, Henderson IR, Polticelli F, Pizza M, Rosini R, Desvaux M. Front Microbiol. 8:2607. Pubmed
- Establishment of Constraints on Amyloid Formation Imposed by Steric Exclusion of Globular Domains. Azizyan RA, Garro A, Radkova Z, Anikeenko A, Bakulina A, Dumas C, Kajava AV. J Mol Biol. 430(20):3835-3846. Pubmed
2017
- Classification of β-hairpin repeat proteins. Roche DB, Viet PD, Bakulina A, Hirsh L, Tosatto SCE, Kajava AV. J Struct Biol. pii: S1047-8477(17)30162-4. Pubmed
- Usage of a dataset of NMR resolved protein structures to test aggregation versus solubility prediction algorithms. Roche DB, Villain E, Kajava AV. Protein Sci. 26(9):1864-1869. Pubmed
- DisProt 7.0: a major update of the database of disordered proteins. Piovesan D, Tabaro F, Mičetić I, Necci M, Quaglia F, Oldfield CJ, Aspromonte MC, Davey NE, Davidović R, Dosztányi Z,[…] Nucleic Acids Res. 45:D219-D227. Pubmed
- RepeatsDB 2.0: improved annotation, classification, search and visualization of repeat protein structures. Paladin L, Hirsh L, Piovesan D, Andrade-Navarro MA, Kajava AV, Tosatto SC. Nucleic Acids Res. 45:D308-D312. Pubmed
2016
- Functional assignment to positively selected sites in the core type III effector RipG7 from Ralstonia solanacearum. Wang K, Remigi P, Anisimova M, Lonjon F, Kars I, Kajava A, Li CH, Cheng CP, Vailleau F, Genin S, Peeters N. Mol Plant Pathol. 17(4):553-64. Pubmed
- Global genetic diversity of the Plasmodium vivax transmission-blocking vaccine candidate Pvs48/45. Vallejo AF, Martinez NL, Tobon A, Alger J, Lacerda MV, Kajava AV, Arévalo-Herrera M, Herrera S. Malar J. 15:202. Pubmed
- Tally: a scoring tool for boundary determination between repetitive and non-repetitive protein sequences. Richard FD, Alves R, Kajava AV. Bioinformatics. 32:1952-8. Pubmed
- α-Synuclein Amyloid Fibrils with Two Entwined, Asymmetrically Associated Protofibrils. Dearborn AD, Wall JS, Cheng N, Heymann JB, Kajava AV, Varkey J, Langen R, Steven AC. J Biol Chem. 291:2310-8. Pubmed
2015
- In search of the boundary between repetitive and non-repetitive protein sequences. Richard FD, Kajava AV. Biochem Soc Trans. 43:807-11. Pubmed
- Restricted mobility of side chains on concave surfaces of solenoid proteins may impart heightened potential for intermolecular interactions. Ramya L, Gautham N, Chaloin L, Kajava AV. Proteins. 83:1654-64. Pubmed
- Single substitution in bacteriophage T4 RNase H alters the ratio between its exo- and endonuclease activities. Kholod N, Sivogrivov D, Latypov O, Mayorov S, Kuznitsyn R, Kajava AV, Shlyapnikov M, Granovsky I. Mutat Res. 781:49-57. Pubmed
- Evolution of proteasome regulators in eukaryotes. Fort P, Kajava AV, Delsuc F, Coux O. Genome Biol Evol. 7:1363-79. Pubmed
- TAPO: A combined method for the identification of tandem repeats in protein structures. Do Viet P, Roche DB, Kajava AV. FEBS Lett. 589:2611-9. Pubmed
- Structure-based view on [PSI(+)] prion properties. Bondarev SA, Zhouravleva GA, Belousov MV, Kajava AV. Prion. 9:190-9. Pubmed
- Designs on a curve. Bazan JF, Kajava AV. Nat Struct Mol Biol. 22:103-5. Pubmed
- A structure-based approach to predict predisposition to amyloidosis. Ahmed AB, Znassi N, Château MT, Kajava AV. Alzheimers Dement. 11:681-90. Pubmed
2014
- TRDistiller: a rapid filter for enrichment of sequence datasets with proteins containing tandem repeats. Richard FD, Kajava AV. J Struct Biol. 186:386-91. Pubmed
- Evolutionary link between metazoan RHIM motif and prion-forming domain of fungal heterokaryon incompatibility factor HET-s/HET-s. Kajava AV, Klopffleisch K, Chen S, Hofmann K. Sci Rep. 4:7436. Pubmed
- Conformational switching in PolyGln amyloid fibrils resulting from a single amino acid insertion. Huang RK1 Baxa U, Aldrian G, Ahmed AB, Wall JS, Mizuno N, Antzutkin O, Steven AC, Kajava AV. Biophys J. 106:2134-42. Pubmed
- RepeatsDB: a database of tandem repeat protein structures. Di Domenico T, Potenza E, Walsh I, Parra RG, Giollo M, Minervini G, Piovesan D, Ihsan A, Ferrari C, Kajava AV, Tosatto SC. Nucleic Acids Res. 42:D352-7. Pubmed
- Plasmodium vivax antigen discovery based on alpha-helical coiled coil protein motif. Céspedes N, Habel C, Lopez-Perez M, Castellanos A, Kajava AV, Servis C, Felger I, Moret R, Arévalo-Herrera M, Corradin G, Herrera S. PLoS One. 9:e100440. Pubmed
- Drosophila Spag is the homolog of RNA polymerase II-associated protein 3 (RPAP3) and recruits the heat shock proteins 70 and 90 (Hsp70 and Hsp90) during the assembly of cellular machineries. Benbahouche Nel H, Iliopoulos I, Török I, Marhold J, Henri J, Kajava AV, Farkaš R, Kempf T, Schnölzer M, Meyer P, Kiss I, Bertrand E, Mechler BM, Pradet-Balade B. J Biol Chem. 289:6236-47. Pubmed
- Plasmodium falciparum merozoite surface protein 2: epitope mapping and fine specificity of human antibody response against non-polymorphic domains. Balam S, Olugbile S, Servis C, Diakité M, D’Alessandro A, Frank G, Moret R, Nebie I, Tanner M, Felger I, Smith T, Kajava AV, Spertini F, Corradin G. Malar J. 13:510. Pubmed
- A structure-based approach to predict predisposition to amyloidosis. Ahmed AB, Znassi N, Château MT, Kajava AV. Alzheimers Dement. 11:681-90. Pubmed
2013
- Breaking the amyloidogenicity code: methods to predict amyloids from amino acid sequence. Ahmed AB, Kajava AV. FEBS Lett. 587(8):1089-95. Pubmed
- Antigenicity and immunogenicity of a novel chimeric peptide antigen based on the P. vivax circumsporozoite protein. Céspedes N, Arévalo-Herrera M, Felger I, Reed S, Kajava AV, Corradin G, Herrera S. Vaccine. 31(42):4923-30. Pubmed
- Effect of charged residues in the N-domain of Sup35 protein on prion [PSI+] stability and propagation. Bondarev SA, Shchepachev VV, Kajava AV, Zhouravleva GA. J Biol Chem. 288(40):28503-13. Pubmed
- EpCAM controls actomyosin contractility and cell adhesion by direct inhibition of PKC. Maghzal N, Kayali HA, Rohani N, Kajava AV, Fagotto F. Dev Cell. 27(3):263-77. Pubmed
2012
- Tandem repeats in proteins: from sequence to structure. Kajava AV. J Struct Biol. 179(3):279-88. Pubmed
- Malaria vaccine development using synthetic peptides as a technical platform. Corradin G, Céspedes N, Verdini A, Kajava AV, Arévalo-Herrera M, Herrera S. Adv Immunol. 114:107-49. Pubmed
- PRDB: Protein Repeat DataBase. Jorda J, Baudrand T, Kajava AV. Proteomics. 12(9):1333-6. Pubmed
- Repeat or not repeat?–Statistical validation of tandem repeat prediction in genomic sequences. Schaper E, Kajava AV, Hauser A, Anisimova M. Nucleic Acids Res. 40(20):10005-17. Pubmed
- Cell biological characterization of the malaria vaccine candidate trophozoite exported protein 1. Kulangara C, Luedin S, Dietz O, Rusch S, Frank G, Mueller D, Moser M, Kajava AV, Corradin G, Beck HP, Felger I. PLoS One. 7(10):e46112. Pubmed
2011
- Identification of residues within tropomodulin-1 responsible for its localization at the pointed ends of the actin filaments in cardiac myocytes. Tsukada T, Kotlyanskaya L, Huynh R, Desai B, Novak SM, Kajava AV, Gregorio CC, Kostyukova AS. J Biol Chem. 286(3):2194-204. Pubmed
- Malaria vaccine candidate: design of a multivalent subunit α-helical coiled coil poly-epitope. Olugbile S, Villard V, Bertholet S, Jafarshad A, Kulangara C, Roussilhon C, Frank G, Agak GW, Felger I, Nebie I, Konate K, Kajava AV, Schuck P, Druilhe P, Spertini F, Corradin G. 29(40):7090-9. Pubmed
2010
- A network of hydrogen bonds on the surface of TLR2 controls ligand positioning and cell signaling. Kajava AV, Vasselon T. J Biol Chem. 2010 285(9):6227-34. Pubmed
- Beta arcades: recurring motifs in naturally occurring and disease-related amyloid fibrils. Kajava AV, Baxa U, Steven AC. FASEB J. 24(5):1311-9. Pubmed
- Malaria vaccine: why is it taking so long? Corradin G, Kajava AV. Expert Rev Vaccines. 9(2):111-4. Pubmed
- Protein tandem repeats – the more perfect, the less structured. Jorda J, Xue B, Uversky VN, Kajava AV. FEBS J. 277(12):2673-82. Pubmed
- Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex. Pérez-Victoria FJ, Abascal-Palacios G, Tascón I, Kajava A, Magadán JG, Pioro EP, Bonifacino JS, Hierro A. Proc Natl Acad Sci U S A. 107(29):12860-5. Pubmed
- Protein homorepeats sequences, structures, evolution, and functions. Jorda J, Kajava AV. Adv Protein Chem Struct Biol. 79:59-88. Pubmed
- Long synthetic peptides for the production of vaccines and drugs: a technological platform coming of age. Corradin G, Kajava AV, Verdini A. Sci Transl Med. 2(50):50rv3. Pubmed
2009
- The C-terminal domain of Plasmodium falciparum merozoite surface protein 3 self-assembles into alpha-helical coiled coil tetramer. Gondeau C, Corradin G, Heitz F, Le Peuch C, Balbo A, Schuck P, Kajava AV. Mol Biochem Parasitol. 165(2):153-61. Pubmed
- Sequence conservation in Plasmodium falciparum alpha-helical coiled coil domains proposed for vaccine development. Kulangara C, Kajava AV, Corradin G, Felger I. PLoS One. 4(5):e5419. Pubmed
- T-REKS: identification of Tandem REpeats in sequences with a K-meanS based algorithm. Jorda J, Kajava AV. Bioinformatics. 25(20):2632-8. Pubmed
- EtpB is a pore-forming outer membrane protein showing TpsB protein features involved in the two-partner secretion system. Meli AC, Kondratova M, Molle V, Coquet L, Kajava AV, Saint N. J Membr Biol. 230(3):143-54. Pubmed
- Vaccine potentials of an intrinsically unstructured fragment derived from the blood stage-associated Plasmodium falciparum protein PFF0165c. Olugbile S, Kulangara C, Bang G, Bertholet S, Suzarte E, Villard V, Frank G, Audran R, Razaname A, Nebie I, Awobusuyi O, Spertini F, Kajava AV, Felger I, Druilhe P, Corradin G. Infect Immun. 77(12):5701-9. Pubmed
2008
- Three-dimensional structure of canine adenovirus serotype 2 capsid. Schoehn G, El Bakkouri M, Fabry CM, Billet O, Estrozi LF, Le L, Curiel DT, Kajava AV, Ruigrok RW, Kremer EJ. J Virol. 82(7):3192-203. Pubmed
- Origin and evolution of GALA-LRR, a new member of the CC-LRR subfamily: from plants to bacteria? Kajava AV, Anisimova M, Peeters N. PLoS One. 3(2):e1694. Pubmed
- Longitudinal analyses of immune responses to Plasmodium falciparum derived peptides corresponding to novel blood stage antigens in coastal Kenya. Agak GW, Bejon P, Fegan G, Gicheru N, Villard V, Kajava AV, Marsh K, Corradin G. Vaccine. 26(16):1963-71. Pubmed
Bioinformatique structurale et modélisation moléculaire

Andrey KAJAVA
Chef d’équipe (Chercheur DR1 CNRS)
Membres de l’équipe
(Chercheur DR1) +33 (0)4 34 35 95 38 |
|
(Maître de conférences) +33 (0)4 34 35 95 39 |
|
(Doctorant) +33 (0)4 34 35 95 39 |
|
(IR-Recherche) +33 (0)4 34 35 95 39 |
|
(Chercheur) +33 (0)4 34 35 9 |
|
(Stagiaire) +33 (0)4 34 35 95 39 |

Contactez notre équipe
Remplacez le nom et l’adresse ci-dessus par celui du membre à contacter
prenom.nom@crbm.cnrs.fr





