Our research
Embryo development is a non-equilibrium process exhibiting emergent behaviors—molecular interactions inside cells irreversibly pattern morphological changes on the animal body through regulating developmental genes. To initiate and control gene expression, transcription factors (TFs) must bind to enhancers and promoters. However, transient and low-affinity interactions between TFs and developmental enhancers appear to be prevalent and even necessary for regulatory specificity in animals ranging from nematodes to mammals. How could these weak interactions reliably produce the precise expression patterns that are necessary to control embryogenesis?
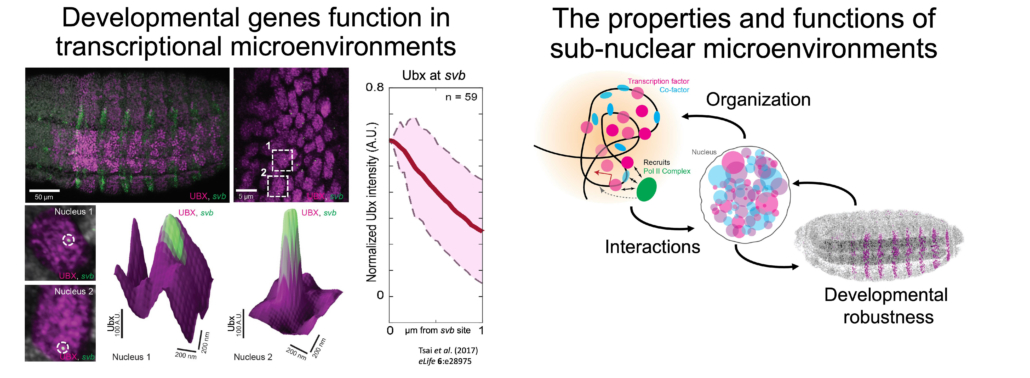
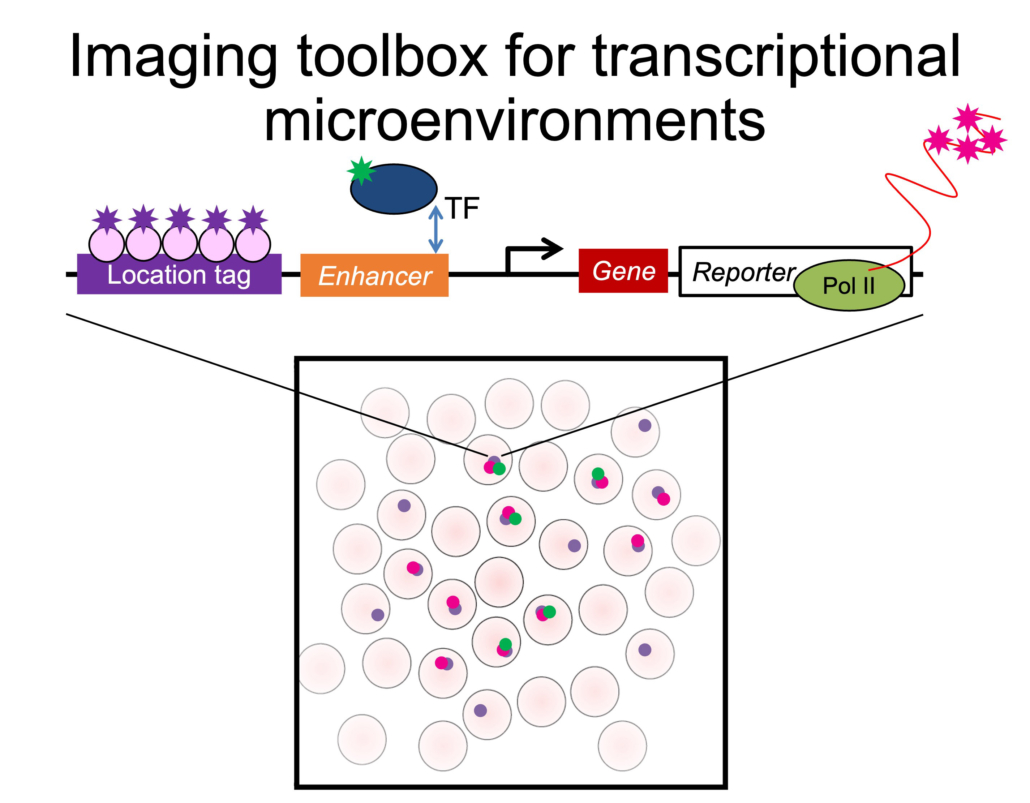
Using Drosophila melanogaster embryos, our work showed that TFs such as the Homeobox (Hox) factor Ultrabithorax (Ubx) coalesce into specialized sub-nuclear microenvironments around the developmental gene shavenbaby (svb). Multiple genes can share and reinforce the same environment, improving the robustness of gene expression and of phenotype development against environmental and genetic perturbations. Thus, the spatial organization of TFs and genes inside the nucleus could be a strategy to facilitate developmental robustness. However, little is known about the physical and interactional properties inside these environments. When do they form during embryogenesis? What determines their location and composition? What roles does the chromatin play? How do genes interact with microenvironments? How do they affect the behaviors of TFs?
To answer these questions, our group combines genetic tools available in Drosophila melanogaster with high- and super-resolution fluorescence imaging to investigate the interaction of genes and TFs in transcriptional microenvironments in the context of embryo development.
1) When do microenvironments form and how does nuclear organization guide cellular differentiation?
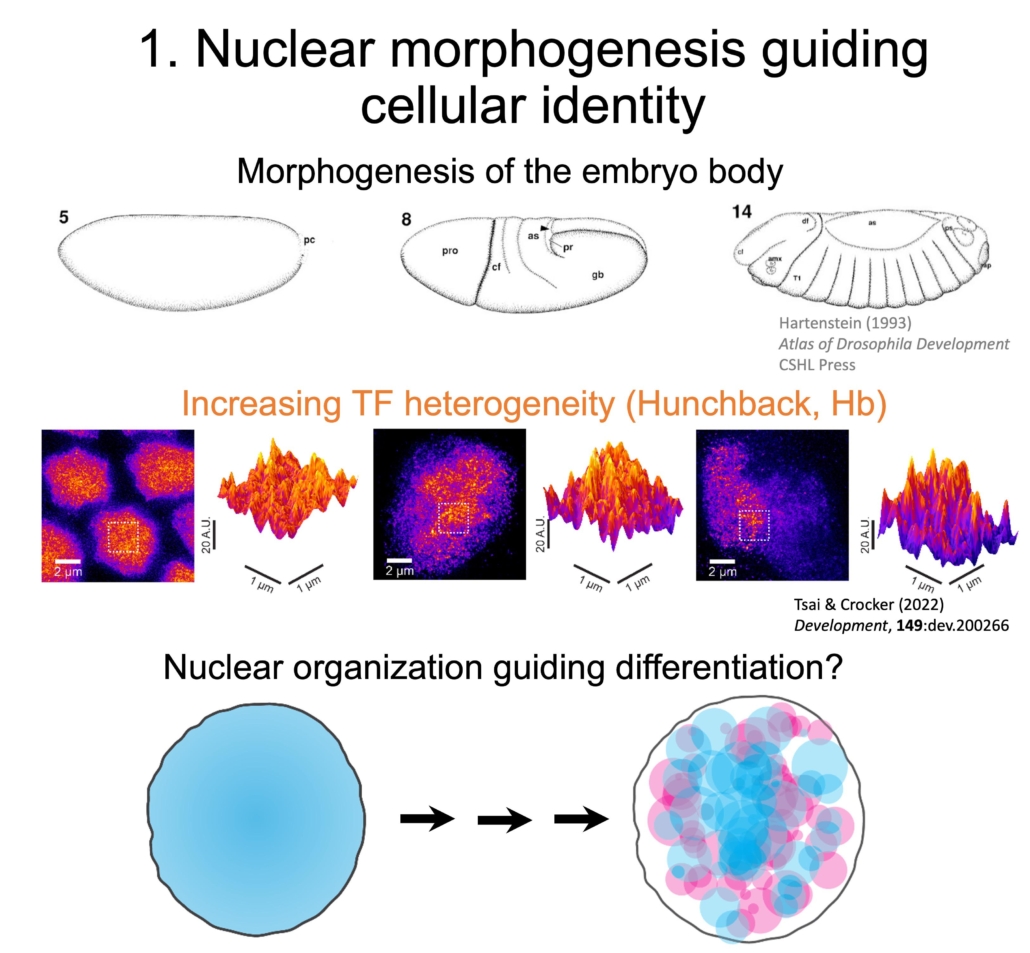
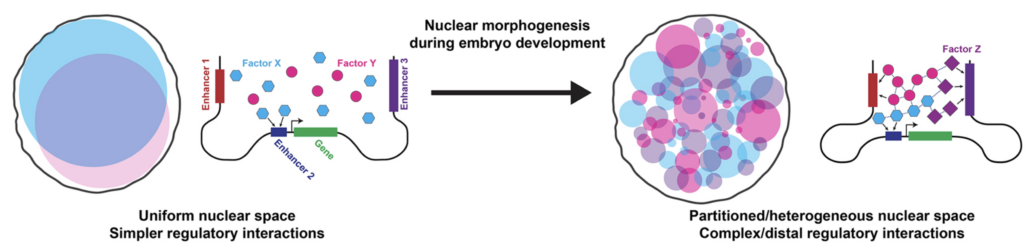
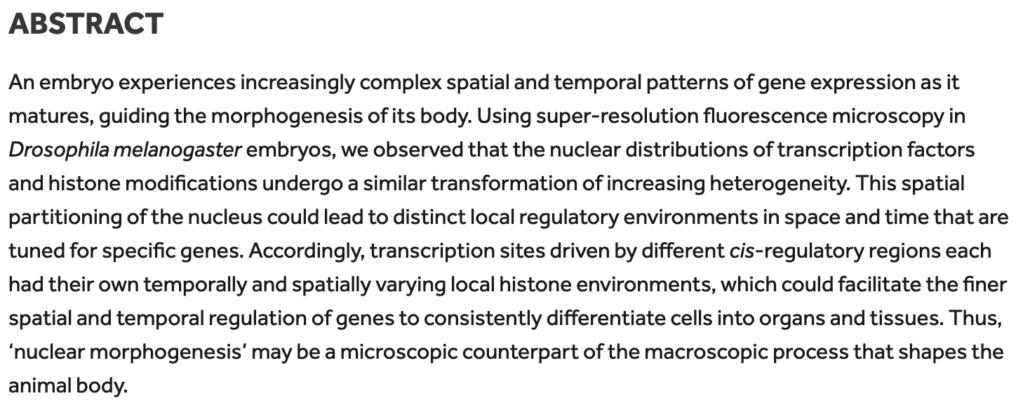
As the embryo matures, its body undergoes morphogenesis, which transforms a fertilized egg into a body with complex structures. We have observed the nuclear distributions of several TFs becoming increasingly heterogeneous during the same time frame. Histone modifications showed a similar increase in heterogeneity as development progresses. This suggests that the nuclear space also undergoes “morphogenesis” that partitions it into distinct compartments.
We are now observing different cell lineages across development to compare how the distributions of TFs and epigenetic marks in the nucleus may guide differentiation.
2) Can colocalization coordinate co-regulation of genes across transcriptional networks?
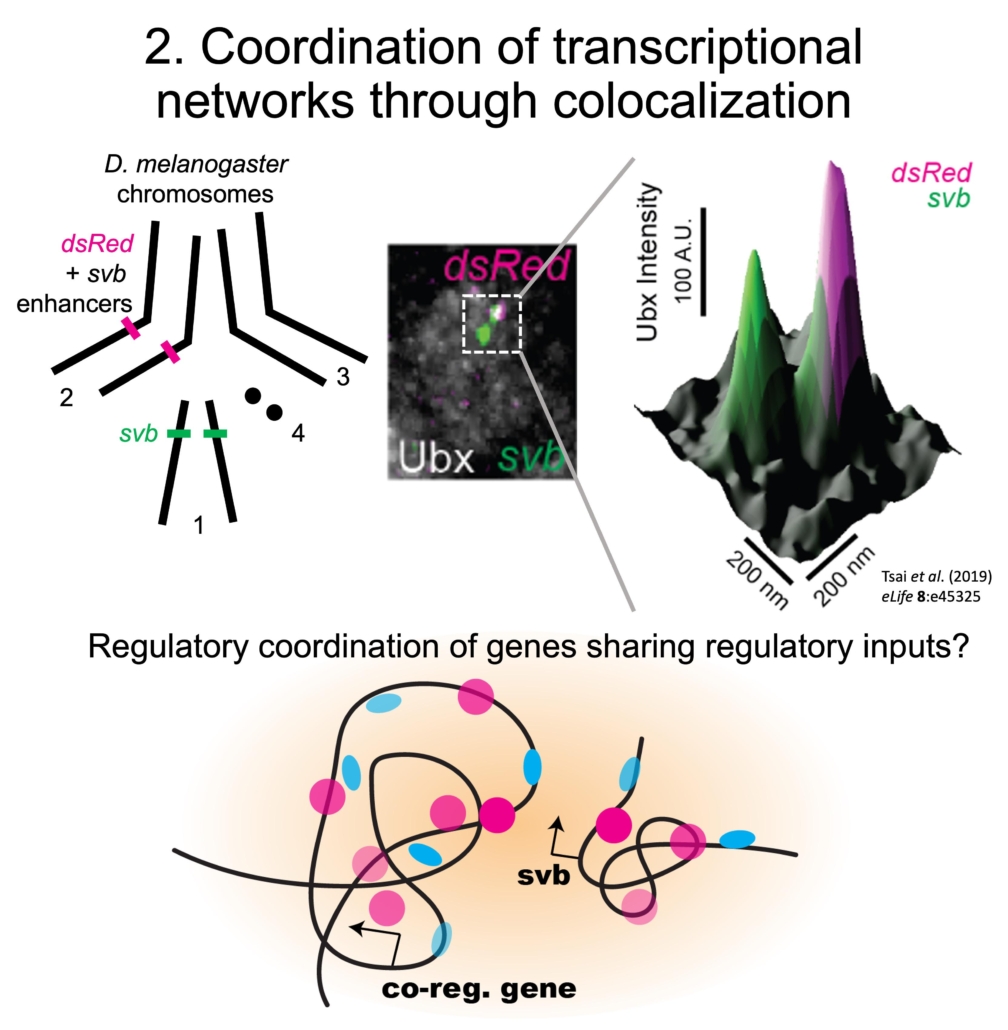
Transgenic transcription sites driven by svb enhancers colocalized with the endogenous svb locus and increased Ubx concentration and svb transcriptional output, ultimately rescuing the trichome phenotype in a fly line with a deficient svb locus. Thus, genes may utilize physical proximity in microenvironments to improve their regulatory coordination and robustness. However, it is not clear if physical proximity of genes and transcriptional activity are related and if colocalization occurs between other genes in transcriptional networks.
Thus, we are developing imaging tools in embryos to monitor the position and transcriptional state of genes to understand the spatial coordination of co-regulated genes in regulatory networks.
3) How do local interactions with proteins and the DNA alter the behaviors of TFs?
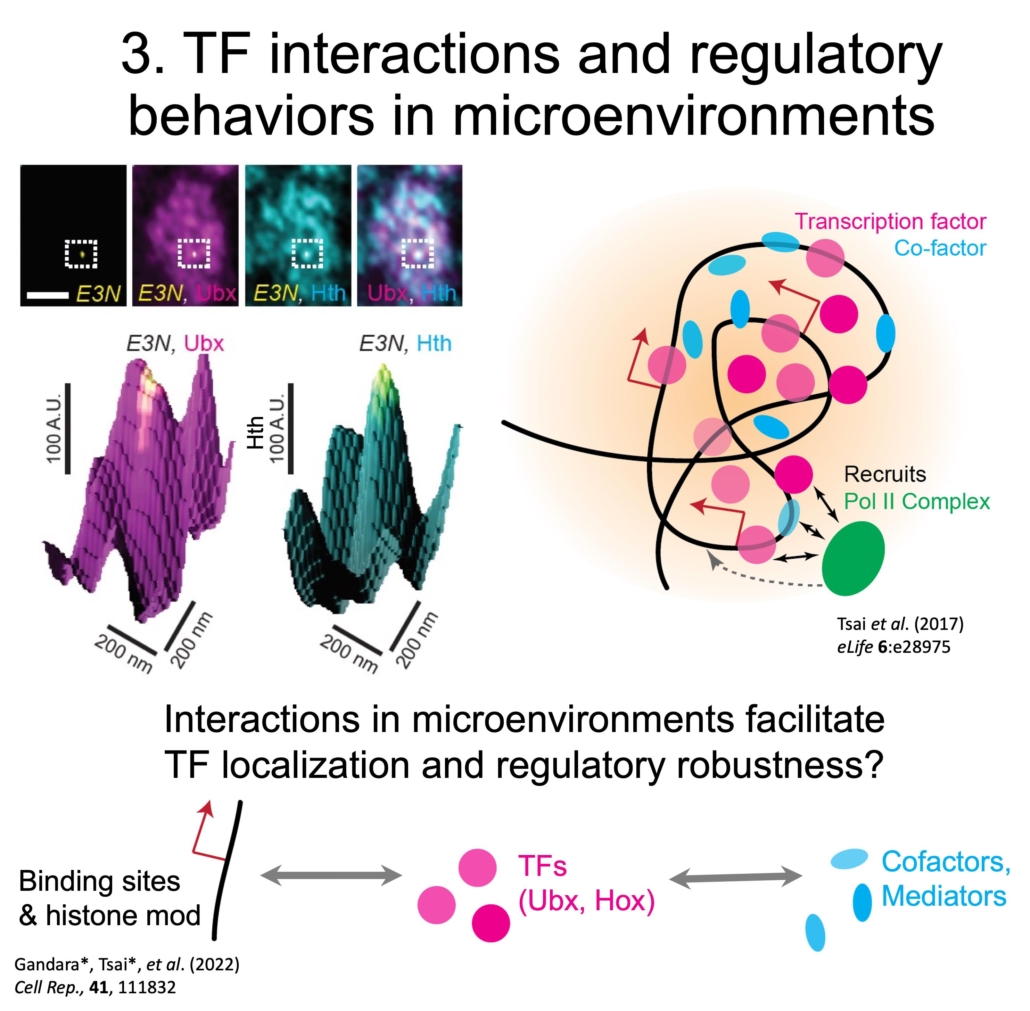
In general, TFs do not colocalize with each other and with markers for transcriptional activity, such as active (S5P) PolII. However, svb transcription sites tended to be enriched for both Ubx and a specific cofactor, Homothorax (Hth). This difference between the global and local overlap of different factors suggests that transcriptional microenvironments are formed using specific interactions. Interestingly, Ubx and its Hox family paralog AbdominalA (AbdA) did not colocalize, even though they have identical consensus DNA binding sequences. This suggests that interactions beyond DNA binding also tune the localization of TFs.
We are therefore tagging developmental TFs for live imaging experiments and modifying their interaction domains to understand how TFs localize and how their regulatory behaviors change based on local TF-DNA and TF-protein interactions in microenvironments.
Read more
Publications
2022
- Developmental phenomics suggests that H3K4 monomethylation confers multi-level phenotypic robustness. Gandara L, Tsai A, Ekelöf M, Galupa R, Preger-Ben Noon E, Alexandrov T, Crocker J. Cell Rep. 2022 Dec 13;41(11):111832. Pubmed
- Nuclear morphogenesis: forming a heterogeneous nucleus during embryogenesis. Tsai A, Crocker J. Development. 2022 Feb 15;149(4):dev200266. Pubmed
2020
- Dense and pleiotropic regulatory information in a developmental enhancer. Fuqua T, Jordan J, van Breugel ME, Halavatyi A, Tischer C, Polidoro P, Abe N, Tsai A, Mann RS, Stern DL, Crocker J. Nature. 2020 Nov;587(7833):235-239. Pubmed
- Robust and efficient gene regulation through localized nuclear microenvironments. Tsai A, Galupa R, Crocker J. Development. 2020 Oct 5;147(19):dev161430. Pubmed
2019
- Multi-enhancer transcriptional hubs confer phenotypic robustness. Tsai A, Alves MR, Crocker J. Elife. 2019 Jul 11;8:e45325. Pubmed
2018
- Visualizing long-range enhancer-promoter interaction. Tsai A, Crocker J. Nat Genet. 2018 Sep;50(9):1205-1206. Pubmed
- Transvection Goes Live-Visualizing Enhancer-Promoter Communication between Chromosomes. Tsai A, Singer RH, Crocker J. Mol Cell. 2018 Apr 19;70(2):195-196. Pubmed
2017
- Nuclear microenvironments modulate transcription from low-affinity enhancers. Tsai A, Muthusamy AK, Alves MR, Lavis LD, Singer RH, Stern DL, Crocker J. Elife. 2017 Nov 2;6:e28975. Pubmed
- A Fully Synthetic Transcriptional Platform for a Multicellular Eukaryote. Crocker J, Tsai A, Stern DL. Cell Rep. 2017 Jan 3;18(1):287-296. Pubmed
2016
- Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Zhang Z, English BP, Grimm JB, Kazane SA, Hu W, Tsai A, Inouye C, You C, Piehler J, Schultz PG, Lavis LD, Revyakin A, Tjian R. Genes Dev. 2016 Sep 15;30(18):2106-2118. Pubmed
- Glutamate-induced RNA localization and translation in neurons. Yoon YJ, Wu B, Buxbaum AR, Das S, Tsai A, English BP, Grimm JB, Lavis LD, Singer RH. Proc Natl Acad Sci U S A. 2016 Nov 1;113(44):E6877-E6886. 35:415-26. Pubmed`
- Amino acid sequence repertoire of the bacterial proteome and the occurrence of untranslatable sequences. Navon SP, Kornberg G, Chen J, Schwartzman T, Tsai A, Puglisi EV, Puglisi JD, Adir N. Proc Natl Acad Sci U S A. 2016 Jun 28;113(26):7166-70. Pubmed
2015
- Probing the Translation Dynamics of Ribosomes Using Zero-Mode Waveguides. Tsai A, Puglisi JD, Uemura S. Prog Mol Biol Transl Sci. 2016;139:1-43. Pubmed
2014
- Dynamic pathways of -1 translational frameshifting. Chen J, Petrov A, Johansson M, Tsai A, O’Leary SE, Puglisi JD. Nature. 2014 Aug 21;512(7514):328-32. Pubmed
- Sequence-dependent elongation dynamics on macrolide-bound ribosomes. Johansson M, Chen J, Tsai A, Kornberg G, Puglisi JD. Cell Rep. 2014 Jun 12;7(5):1534-1546. Pubmed
- The dynamics of SecM-induced translational stalling. Tsai A, Kornberg G, Johansson M, Chen J, Puglisi JD. Cell Rep. 2014 Jun 12;7(5):1521-1533. Pubmed
- Signal recognition particle-ribosome binding is sensitive to nascent chain length. Noriega TR, Tsai A, Elvekrog MM, Petrov A, Neher SB, Chen J, Bradshaw N, Puglisi JD, Walter P. J Biol Chem. 2014 Jul 11;289(28):19294-305. Pubmed
- High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Chen J, Dalal RV, Petrov AN, Tsai A, O’Leary SE, Chapin K, Cheng J, Ewan M, Hsiung PL, Lundquist P, Turner SW, Hsu DR, Puglisi JD. Proc Natl Acad Sci U S A. 2014 Jan 14;111(2):664-9. Pubmed
2013
- Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Simonetti A, Marzi S, Billas IM, Tsai A, Fabbretti A, Myasnikov AG, Roblin P, Vaiana AC, Hazemann I, Eiler D, Steitz TA, Puglisi JD, Gualerzi CO, Klaholz BP. Proc Natl Acad Sci U S A. 2013 Sep 24;110(39):15656-61. Pubmed
- Coordinated conformational and compositional dynamics drive ribosome translocation. Chen J, Petrov A, Tsai A, O’Leary SE, Puglisi JD. Nat Struct Mol Biol. 2013 Jun;20(6):718-27. Pubmed
- The impact of aminoglycosides on the dynamics of translation elongation. Tsai A, Uemura S, Johansson M, Puglisi EV, Marshall RA, Aitken CE, Korlach J, Ehrenberg M, Puglisi JD. Cell Rep. 2013 Feb 21;3(2):497-508. Pubmed
2012
- Unraveling the dynamics of ribosome translocation. Chen J, Tsai A, O’Leary SE, Petrov A, Puglisi JD. Curr Opin Struct Biol. 2012 Dec;22(6):804-14. Pubmed
- Single-molecule analysis of translational dynamics. Petrov A, Chen J, O’Leary S, Tsai A, Puglisi JD. Cold Spring Harb Perspect Biol. 2012 Sep 1;4(9):a011551. Pubmed
- Isolation of NEDDylated proteins in human cells. Leidecker O, Xirodimas DP. Methods Mol Biol. 832:133-40. Pubmed
- Heterogeneous pathways and timing of factor departure during translation initiation. Tsai A, Petrov A, Marshall RA, Korlach J, Uemura S, Puglisi JD. Nature. 2012 Jul 19;487(7407):390-3. Pubmed
- Chen J, Tsai A, Petrov A, Puglisi JD. Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics. J Am Chem Soc. 2012 Apr 4;134(13):5734-7. Pubmed
2011
- Petrov A, Kornberg G, O’Leary S, Tsai A, Uemura S, Puglisi JD. Dynamics of the translational machinery. Curr Opin Struct Biol. 2011 Feb;21(1):137-45. Pubmed
Nuclear organization of transcription in embryos
 Albert TSAI
Albert TSAI
Group leader (Researcher CNRS)
Team Members
(Chercheur CRCN) +33 (0)4 34 35 95 05 |
|
(Doctorant) +33 (0)4 34 35 95 01 |
|
(Chercheur) +33 (0)4 34 35 95 01 |
|
(Stagiaire) +33 (0)4 34 35 9 |

Contact us
Replace the name and address below with that of the member to contact
Firstname.name@crbm.cnrs.fr