Our Research
Asymmetric cell division is the main mode of division of stem cells and plays a central role in generating cell diversity during development. Indeed, during asymmetric cell division of polarized cells, biomolecules are asymmetrically segregated between daughter cells. The position of the mitotic spindle within the cell determines whether the cell will divide symmetrically or asymmetrically. Spindle positioning, in turn, depends on the interaction of astral microtubules (cMTs) with cortical factors. A complex network of proteins, such as non-motor microtubule-associated proteins (plus-end tracking proteins, +TIPs), kinesins, dynein and actin-interacting proteins, are involved in these interactions.
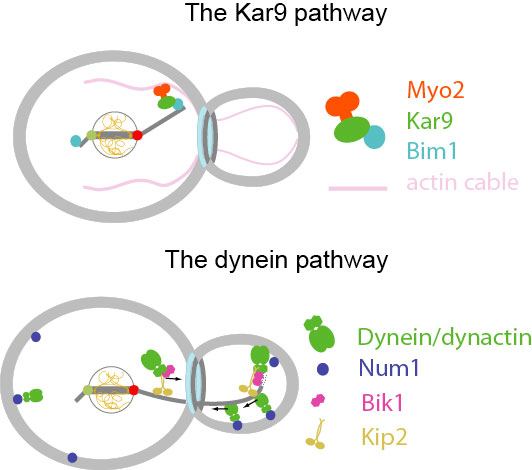
Budding yeast also divides asymmetrically and is used as a model organism to study asymmetric cell division. Spindle positioning in yeast depends on two genetically identified pathways: i) the dynein pathway that includes complexes of the microtubule-dependent motor dynein, and ii) the Kar9 pathway. Kar9 is the yeast functional equivalent of the Adenomatous Polyposis Coli (APC) tumour suppressor, a protein with a central role in spindle positioning from Drosophila to mammals. Dynein, Kar9 and APC homologues are universal mediators of spindle positioning during eukaryotic asymmetric division, but their regulation is poorly understood.
In the laboratory, we explore how cell cycle regulators modulate the temporal activation and dynamics of dynein and Kar9 complexes during spindle positioning in budding yeast. During their transport along microtubules and interactions with cortical proteins, these factors form highly dynamic protein complexes. Cyclin-dependent kinases control both the formation of these complexes and the coordination of their activity with other cytoskeletal events, such as chromosome segregation and cytokinesis. To understand the regulation of Kar9 and dynein complexes we use genetics to study their interactions. Using protein biochemistry, we analyse the Kar9/dynein complex composition and phosphorylation. Finally, we investigate complex dynamics in living yeast cells by time-lapse fluorescence imaging.
En savoir plus
Financements
ANR (Didier Portran)
Publications
2025
- The kinesin Kip2 promotes microtubule growth using a bipartite polymerase module to deliver tubulin to microtubule plus ends. Nadalis S, Neyret A, Xanthou I, Siaden-Ortega R, Marcoux J, Abrieu A, Drechsler H, Liakopoulos D, Portran D. Cell Rep. 2025 Sep 27;44(10):116353. Pubmed
2023
- The kinesin Kip2 promotes microtubule growth using a bipartite polymerase module to deliver tubulin to microtubule plus ends. Nadalis S, Neyret A, Xanthou I, Siaden-Ortega R, Marcoux J, Abrieu A, Drechsler H, Liakopoulos D, Portran D. Cell Rep. 2025 Sep 27;44(10):116353. Pubmed
- The motor domain of the kinesin Kip2 promotes microtubule polymerization at microtubule tipsThe kinesin Kip2 promotes microtubule growth using a bipartite polymerase module to deliver tubulin to microtubule plus ends.. Chen X, Portran D, Widmer LA, Stangier MM, Czub MP, Liakopoulos D, Stelling J, Steinmetz MO, Barral Y. J Cell Biol. 2023 Jul 3;222(7):e202110126. Pubmed
2021
- Coupling DNA Replication and Spindle Function in Saccharomyces cerevisiae. Liakopoulos D. Cells. 2021 Nov 30;10(12):3359. Pubmed
2019
- How Does SUMO Participate in Spindle Organization? Abrieu A, Liakopoulos D. Cells. 8(8). Pubmed
- Lattice defects induce microtubule self-renewal. Schaedel L, Triclin S, Chrétien D, Abrieu A, Aumeier C, Gaillard J, Blanchoin L, Théry M, John K.. Nat Phys. 2019 Aug;15(8):830-838. Pubmed
2018
- An E2-ubiquitin thioester-driven approach to identify substrates modified with ubiquitin and ubiquitin-like molecules. Bakos G, Yu L, Gak IA, Roumeliotis TI, Liakopoulos D, Choudhary JS, Mansfeld J. Nat Commun. 9(1):4776. Pubmed
2016
- Kar9 controls the nucleocytoplasmic distribution of yeast EB1. J. Schweiggert, D. Panigada, AN. Tan, D. Liakopoulos Cell Cycle, 15 : 2860-2866. Pubmed
- Regulation of a Spindle Positioning Factor at Kinetochores by SUMO-Targeted Ubiquitin Ligases. Schweiggert J, Stevermann L, Panigada D, Kammerer D, Liakopoulos D. Dev Cell. 36:415-27. Pubmed
2015
- Kinesin Kip2 enhances microtubule growth in vitro through length-dependent feedback on polymerization and catastrophe. Hibbel A, Bogdanova A, Mahamdeh M, Jannasch A, Storch M, Schäffer E, Liakopoulos D, Howard J. Elife. 4: e10542. Pubmed
2013
- An auxiliary, membrane-based mechanism for nuclear migration in budding yeast. Kirchenbauer M, Liakopoulos D. Mol Biol Cell. 24(9):1434-43. Pubmed
2010
- Ubiquitylation regulates interactions of astral microtubules with the cleavage apparatus. Kammerer D, Stevermann L, Liakopoulos D. Curr Biol. 20(14):1233-43. Pubmed
- Arp1, an actin-related protein, in Plasmodium berghei. Siden-Kiamos I, Schüler H, Liakopoulos D, Louis C. Mol Biochem Parasitol. 173(2):88-96. Pubmed
2008
- Regulation of mitotic spindle asymmetry by SUMO and the spindle-assembly checkpoint in yeast. Leisner C, Kammerer D, Denoth A, Britschi M, Barral Y, Liakopoulos D. Curr Biol. 18(16):1249-55. Pubmed
Spindle positioning and organization

Dimitris LIAKOPOULOS
Group Leader (Research Director CNRS)
Team Members
(Chercheur DR2) +33 (0)4 34 35 95 36 |
|
(Chercheur DR2) +33 (0)4 34 35 95 67 |
|
(IE-Recherche) +33 (0)4 34 35 95 67 |
|
(CRCN) +33 (0)4 34 35 96 43 |
|
(Doctorant) +33 (0)4 34 35 95 67 |
|
(Stagiaire) +33 (0)4 34 35 9 |

Contact us
Replace the name and address below with that of the member to contact
firstname.name@crbm.cnrs.fr
FINANCIAL SUPPORT