Our Research
The Protein Quality Control (PQC) system
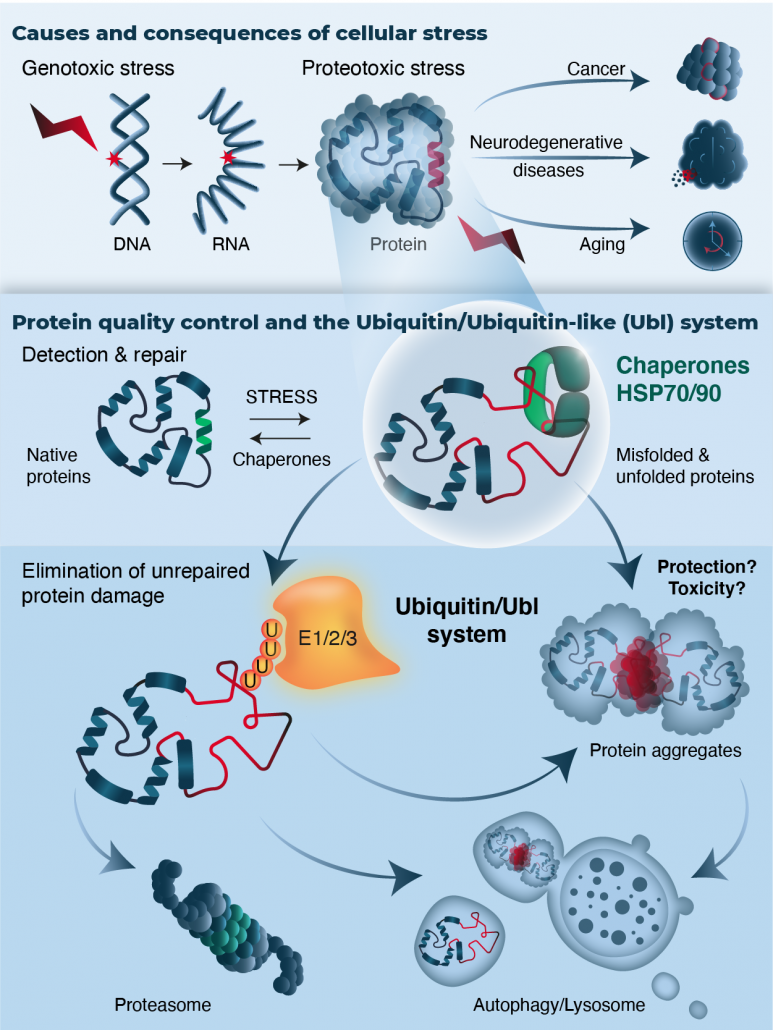
Understanding how organisms respond to environmental stress has critical implications both on quality of life and treatment of diseases. Maintaining a functioning proteome is challenging for cells given the variety of proteotoxic stresses (exposure to chemicals, elevated temperature, oxidative species) that constantly cause protein damage.
Misfolded proteins are deleterious to the cell and this is due either to the loss of their function and/or to their ability to form insoluble aggregates. The generation of aberrant protein aggregates is seen during ageing and has been associated with multiple pathologies known as “conformational disorders”. These include lysosomal storage diseases, cystic fibrosis, cancer and many neurodegenerative diseases such as Alzheimer, Huntington’s, Parkinson’s, Amyotrophic Lateral Sclerosis (ALS).
The maintenance of a healthy proteome (proteostasis) is thus one of the biggest challenges that cells encounter. To counteract and prevent protein misfolding/aggregation, cells have evolved a Protein Quality Control (PQC) network, which is based on molecular chaperones and proteolytic machineries (26S proteasome, lysosomal pathway) that cooperate to maintain proteostasis and thus cellular fitness. The system ensures protein repair and refolding or clearance of terminally damaged proteins.
An integral part of the PQC network is the spatial management of misfolded proteins in the cytoplasm and the nucleus. The sequestration of misfolded proteins to particular subcellular compartments facilitates the removal of damaged proteins. This potentially assists in cell recovery once the stress is alleviated.
Ubiquitin and Ubiquitin-like molecules as key regulators of the PQC system
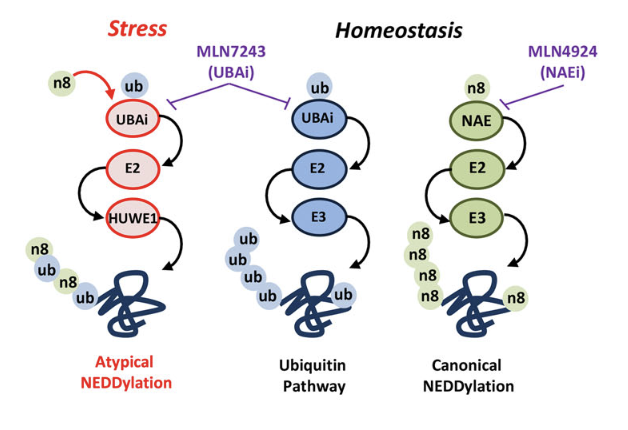
The family of Ubiquitin and Ubiquitin-like molecules (Ubls) such as SUMO and NEDD8, play a critical role in all aspects of the PQC system. The elimination of most aberrant proteins is mediated by the Ubiquitin Proteasome System (UPS) where proteins are marked for degradation via the “Ubiquitination process”. The family of Ubls emerge as important regulators of Ubiquitin signalling and of the PQC system.
Our studies have revealed distinct roles for the Ubiquitin-like molecule NEDD8 as sensor of proteotoxic stress. In particular, we defined the formation of poly-NEDD8 chains and hybrid NEDD8-Ubiquitin chains as molecular signals that specifically operate in the cytoplasm and the nucleus respectively to control the formation of protein inclusions. Our goal is to decipher the role of NEDD8 as regulator of degradation of misfolded proteins and formation of protein aggregates. We combine biochemical, biological, live-imaging, quantitative proteomics in human cells and C.elegans to address the above biological questions. As the NEDD8 pathway is a promising therapeutic target, we envision our studies will reveal strategies for the use of NEDD8 inhibitors in the clinic and potentially to identify new targets within the PQC system for therapeutic intervention.
Read more
Funding
ANR PROCONUC (01/10/2021 – 31/05/2026)
FRM (2026)
LIGUE (2026)
Publications
2025
- SUMOylation of the lysine-less tumor suppressor p14ARF counters ubiquitylation-dependent degradation.El Motiam A, Bouzaher YH, Chen H, Seoane R, Vidal S, Blanquer M, Tolosa RM, Rodríguez-Lemus B, Herrera-Gavilán JA, Vidal A, Palmero I, Rodríguez MS, Sutherland JD, Barrio R, Xirodimas D, Collado M, Bremner R, Rivas C. Cell Death Dis. 2025 Jul 12;16(1):519. Pubmed
- SUMO2/3 conjugation of TDP-43 protects against aggregation. Verde EM, Antoniani F, Mediani L, Secco V, Crotti S, Ferrara MC, Vinet J, Sergeeva A, Yan X, Hoege C, Stuani C, Paron F, Kao TT, Shrivastava R, Polanowska J, Bailly A, Rosa A, Aronica E, Goswami A, Shneider N, Hyman AA, Buratti E, Xirodimas D, Franzmann TM, Alberti S, Carra S. Sci Adv. 2025 Feb 21;11(8):eadq2475. Pubmed
- A nuclear protein quality control system for elimination of nucleolus-related inclusions. Brunello L, Polanowska J, Le Tareau L, Maghames C, Georget V, Guette C, Chaoui K, Balor S, O’Donohue MF, Bousquet MP, Gleizes PE, Xirodimas DP. EMBO J. 2025 Feb;44(3):801-823. Pubmed
2024
- An Anti-Invasive Role for Mdmx through the RhoA GTPase under the Control of the NEDD8 Pathway. Bou Malhab LJ, Schmidt S, Fagotto-Kaufmann C, Pion E, Gadea G, Roux P, Fagotto F, Debant A, Xirodimas DP. Cells. 2024 Sep 28;13(19):1625 Pubmed
- An Anti-Invasive Role for Mdmx through the RhoA GTPase under the Control of the NEDD8 Pathway. Bou Malhab LJ, Schmidt S, Fagotto-Kaufmann C, Pion E, Gadea G, Roux P, Fagotto F, Debant A, Xirodimas DP. Cells. 2024 Sep 28;13(19):1625 Pubmed
- Neddylation inhibition prevents acetaminophen-induced liver damage by enhancing the anabolic cardiolipin pathway.. Gil-Pitarch C, Serrano-Maciá M, Simon J, Mosca L, Conter C, Rejano-Gordillo CM, Zapata-Pavas LE, Peña-Sanfélix P, Azkargorta M, Rodríguez-Agudo R, Lachiondo-Ortega S, Mercado-Gómez M, Delgado TC, Porcelli M, Aurrekoetxea I, Sutherland JD, Barrio R, Xirodimas D, Aspichueta P, Elortza F, Martínez-Cruz LA, Nogueiras R, Iruzubieta P, Crespo J, Masson S, McCain MV, Reeves HL, Andrade RJ, Lucena MI, Mayor U, Goikoetxea-Usandizaga N, González-Recio I, Martínez-Chantar ML. Cell Rep Med. 2024 Jul 16;5(7):101653. Pubmed
- SUMOylation modulates eIF5A activities in both yeast and pancreatic ductal adenocarcinoma cells. Seoane R, Lama-Díaz T, Romero AM, El Motiam A, Martínez-Férriz A, Vidal S, Bouzaher YH, Blanquer M, Tolosa RM, Castillo Mewa J, Rodríguez MS, García-Sastre A, Xirodimas D, Sutherland JD, Barrio R, Alepuz P, Blanco MG, Farràs R, Rivas C. Cell Mol Biol Lett. 2024 Jan 16;29(1):15. Pubmed
2023
- Two distinct mechanisms lead to either oocyte or spermatocyte decrease in C. elegans after whole developmental exposure to γ-rays. Dufourcq Sekatcheff E, Godon C, Bailly A, Quevarec L, Camilleri V, Galas S, Frelon S. PLoS One. 2023 Nov 27;18(11):e0294766. Pubmed
- Deneddylation of ribosomal proteins promotes synergy between MLN4924 and chemotherapy to elicit complete therapeutic responses. Aubry A, Pearson JD, Charish J, Yu T, Sivak JM, Xirodimas DP, Avet-Loiseau H, Corre J, Monnier PP, Bremner R. Cell Rep. 2023 Aug 7;42(8):112925. Pubmed
- Targeting the NEDP1 enzyme to ameliorate ALS phenotypes through stress granule disassembly. Kassouf T, Shrivastava R, Meszka I, Bailly A, Polanowska J, Trauchessec H, Mandrioli J, Carra S, Xirodimas DP. Sci Adv. 2023 Mar 31;9(13):eabq7585. Pubmed
- A Mass Spectrometry-Based Strategy for Mapping Modification Sites for the Ubiquitin-Like Modifier NEDD8. Oliveira CAB, Isaakova E, Beli P, Xirodimas DP. Methods Mol Biol. 2023;2602:137-149. Pubmed
2022
- Mixed in chains: NEDD8 polymers in the Protein Quality Control system. Meszka I, Polanowska J, Xirodimas DP. Semin Cell Dev Biol. 2022 Dec;132:27-37. Pubmed
2021
- Neddylation inhibition ameliorates steatosis in NAFLD by boosting hepatic fatty acid oxidation via the DEPTOR-mTOR axis. Serrano-Maciá M, Simón J, González-Rellan MJ, Azkargorta M, Goikoetxea-Usandizaga N, Lopitz-Otsoa F, De Urturi DS, Rodríguez-Agudo R, Lachiondo-Ortega S, Mercado-Gomez M, Gutiérrez de Juan V, Bizkarguenaga M, Fernández-Ramos D, Buque X, Baselli GA, Valenti LVC, Iruzubieta P, Crespo J, Villa E, Banales JM, Avila MA, Marin JJG, Aspichueta P, Sutherland J, Barrio R, Mayor U, Elortza F, Xirodimas DP, Nogueiras R, Delgado TC, Martínez-Chantar ML. Mol Metab. 101275 Pubmed
- The HSP70 chaperone as sensor of the NEDD8 cycle upon DNA damage. Bailly AP, Xirodimas DP. Biochem Soc Trans. 49(3):1075-1083. Pubmed
- Proteome-wide identification of NEDD8 modification sites reveals distinct proteomes for canonical and atypical NEDDylation. Sofia Lobato-Gil, Jan B. Heidelberger, Chantal Maghames, Aymeric Bailly, Lorene Brunello, Manuel S. Rodriguez, Petra Beli, and Dimitris P. Xirodimas. Cell Rep. 34(3):108635. Pubmed
2020
- Development of Nanobodies as first-in-class inhibitors for the NEDP1 deNEDDylating enzyme. Naima Abidi, Helene Trauchessec, Gholamreza Hassanzadeh-Ghassabeh, Martine Pugniere, Serge Muyldermans, Dimitris P. Xirodimas. bioRxiv 2020.03.20.999326 Pubmed
- Antibody RING-Mediated Destruction of Endogenous Proteins. Ibrahim AFM, Shen L, Tatham MH, Dickerson D, Prescott AR, Abidi N, Xirodimas DP, Hay RT. Mol Cell ; S1097-2765(20)30274-4. Pubmed
2019
- The balance between Mono- and NEDD8-Chains Controlled by NEDP1 upon DNA Damage Is a Regulatory Module of the HSP70 ATPase Activity. Bailly AP, Perrin A, Serrano-Macia M, Maghames C, Leidecker O, Trauchessec H, Martinez-Chantar ML, Gartner A, Xirodimas DP. Cell Rep. 29(1):212-224 Pubmed
2018
- NEDDylation promotes nuclear protein aggregation and protects the Ubiquitin Proteasome System upon proteotoxic stress. Maghames CM, Lobato-Gil S, Perrin A, Trauchessec H, Rodriguez MS, Urbach S, Marin P, Xirodimas DP. Nat Commun. 9(1):4376. Pubmed
- Interplay between SUMOylation and NEDDylation regulates RPL11 localization and function. El Motiam A, Vidal S, de la Cruz-Herrera CF, Da Silva-Álvarez S, Baz-Martínez M, Seoane R, Vidal A, Rodríguez MS, Xirodimas DP, Carvalho AS, Beck HC, Matthiesen R, Collado M, Rivas C. FASEB J. :fj201800341RR Pubmed
2017
- Quantitative FLIM-FRET Microscopy to Monitor Nanoscale Chromatin Compaction In Vivo Reveals Structural Roles of Condensin Complexes. Llères D, Bailly AP, Perrin A, Norman DG, Xirodimas DP, Feil R. Cell Rep. 18:1791-1803. Pubmed
2016
- The use of the NEDD8 inhibitor MLN4924 (Pevonedistat) in a cyclotherapy approach to protect wild-type p53 cells from MLN4924 induced toxicity. Malhab LJ, Descamps S, Delaval B, Xirodimas DP. Sci Rep. 6:37775. Pubmed
- The NEDD8 inhibitor MLN4924 increases the size of the nucleolus and activates p53 through the ribosomal-Mdm2 pathway. Bailly A, Perrin A, Bou Malhab LJ, Pion E, Larance M, Nagala M, Smith P, O’Donohue MF, Gleizes PE, Zomerdijk J, Lamond AI, Xirodimas DP. 35:415-26. Pubmed
2015
- PKA-dependent phosphorylation of ribosomal protein S6 does not correlate with translation efficiency in striatonigral and striatopallidal medium-sized spiny neurons. Biever A, Puighermanal E, Nishi A, David A, Panciatici C, Longueville S, Xirodimas D, Gangarossa G, Meyuhas O, Hervé D, Girault JA, Valjent E. J Neurosci. 35:4113-30. Pubmed
- Regulation of cancer-related pathways by protein NEDDylation and strategies for the use of NEDD8 inhibitors in the clinic. Abidi N, Xirodimas DP. Endocr Relat Cancer. 22:T55-70. Pubmed
2013
- DNA-binding regulates site-specific ubiquitination of IRF-1. Landré V, Pion E, Narayan V, Xirodimas DP, Ball KL. Biochem J. 449(3):707-17. Pubmed
- DUBs « found in translation »: USP15 controls stability of newly synthesized REST. Xirodimas DP. Cell Cycle. 12(16):2536-7. Pubmed
2012
- Characterization of MRFAP1 turnover and interactions downstream of the NEDD8 pathway. Larance M, Kirkwood KJ, Xirodimas DP, Lundberg E, Uhlen M, Lamond AI. Mol Cell Proteomics. 11(3):M111.014407. Pubmed
- Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Embade N, Fernández-Ramos D, Varela-Rey M, Beraza N, Sini M, Gutiérrez de Juan V, Woodhoo A, Martínez-López N, Rodríguez-Iruretagoyena B, Bustamante FJ, de la Hoz AB, Carracedo A, Xirodimas DP, Rodríguez MS, Lu SC, Mato JM, Martínez-Chantar ML. 55(4):1237-48. Pubmed
- Isolation of NEDDylated proteins in human cells. Leidecker O, Xirodimas DP. Methods Mol Biol. 832:133-40. Pubmed
- The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Leidecker O, Matic I, Mahata B, Pion E, Xirodimas DP. Cell Cycle. 11(6):1142-50. Pubmed
- The p53 isoforms are differentially modified by Mdm2. Camus S, Ménendez S, Fernandes K, Kua N, Liu G, Xirodimas DP, Lane DP, Bourdon JC. Cell Cycle.11(8):1646-55. Pubmed
- Influence of the nuclear membrane, active transport, and cell shape on the Hes1 and p53-Mdm2 pathways: insights from spatio-temporal modelling. Sturrock M, Terry AJ, Xirodimas DP, Thompson AM, Chaplain MA. Bull Math Biol. 74(7):1531-79. Pubmed
2011
- Spatio-temporal modelling of the Hes1 and p53-Mdm2 intracellular signalling pathways. Sturrock M, Terry AJ, Xirodimas DP, Thompson AM, Chaplain MA. J Theor Biol. 273(1):15-31. Pubmed
- The essential functions of NEDD8 are mediated via distinct surface regions, and not by polyneddylation in Schizosaccharomyces pombe. Girdwood D, Xirodimas DP, Gordon C. PLoS One. 6(5):e20089. Pubmed
- In the family with ubiquitin. Alexandru G, Pariente N, Xirodimas D. EMBO Rep. 12(9):880-2. Pubmed
- Stable-isotope labeling with amino acids in nematodes. Larance M, Bailly AP, Pourkarimi E, Hay RT, Buchanan G, Coulthurst S, Xirodimas DP, Gartner A, Lamond AI. Nat Methods. 8(10):849-51. Pubmed
2010
- Perturbation of 60 S ribosomal biogenesis results in ribosomal protein L5- and L11-dependent p53 activation. Sun XX, Wang YG, Xirodimas DP, Dai MS. J Biol Chem. 285(33):25812-21. Pubmed
- Mechanism of hypoxia-induced NF-kappaB. Culver C, Sundqvist A, Mudie S, Melvin A, Xirodimas D, Rocha S. Mol Cell Biol. 30(20):4901-21. Pubmed
- Ubiquitin Family Members in the Regulation of the Tumor Suppressor p53. Xirodimas DP, Scheffner M. Subcell Biochem. 54:116-35. Pubmed
2009
- Regulation of nucleolar signalling to p53 through NEDDylation of L11. Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. EMBO Rep. 10(10):1132-9. Pubmed
- Detection of protein SUMOylation in vivo. Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Nat Protoc. 4(9):1363-71. Pubmed
2008
- Ribosomal proteins are targets for the NEDD8 pathway. Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT. EMBO Rep. 9(3):280-6. Pubmed
- Much to know about proteolysis: intricate proteolytic machineries compromise essential cellular functions. Marfany G, Farràs R, Salido E, Xirodimas DP, Rodríguez MS. Biochem Soc Trans. 36(Pt 5):781-5. Pubmed
- Novel substrates and functions for the ubiquitin-like molecule NEDD8. Xirodimas DP. Biochem Soc Trans. 36(Pt 5):802-6. Pubmed
Ubiquitin/Ubiquitin-like molecules and Protein Quality Control
 Dimitris XIRODIMAS
Dimitris XIRODIMAS
Group Leader (Research Director INSERM)
Team Members
(AI-Recherche) +33 (0)4 34 35 95 33 |
|
(Chercheur DR2) +33 (0)4 34 35 95 33 |
|
(CRCN) +33 (0)4 34 35 95 34 |
|
(CRCN) +33 (0)4 34 35 95 33 |
|
(Doctorant) +33 (0)4 34 35 95 33 |
|
(Doctorant) +33 (0)4 34 35 95 33 |

Contact us
Replace the name and address below with that of the member to contact
Firstname.name@crbm.cnrs.fr
Grants