Our Research
For more information please visit our lab website.
Our overall goal is to identify the principles governing the assembly of chromatin regulatory complexes and to characterize their structural organization, activities and regulation by signaling cues. For this, we study the SAGA and TIP60 complexes, which carry histone acetylation, de-ubiquitination and histone variant deposition activities. Our work combines standard molecular, biochemical, and genetic assays with innovative genomic, proteomic, and structural biology approaches, using both fission yeast and cancer cell lines as experimental systems.
Gene regulation is crucial to generate phenotypic diversity between cells of identical genotypes, for example during cell type specification or in response to environmental changes. A critical step in gene expression is transcription, which is controlled by many distinct epigenetic regulators, such as chromatin-modifying and –remodeling complexes. Alterations of their functions have been documented in various human diseases, most notably during neurodegeneration or tumorigenesis. Several chromatin regulatory complexes function as co-activators during transcription initiation, bridging promoter-bound activators to the general transcription machinery. Co-activators have the ability to bind multiple transcription factors and thereby to coordinately regulate the expression of many genes in response to signaling cues.
Despite many studies over the past two decades, several challenges remain in our understanding of co-activator complexes. Our research is articulated around three unanswered questions:
Aim 1: How do signaling pathways control the regulatory activities of transcription co-activators?
Aim 2: How do co-activators sustain both basal transcription and the dynamic regulation of transcription in response to signaling cues?
Aim 3: What are the principles governing co-activator complex assembly and can their assembly be modulated to control their activities?
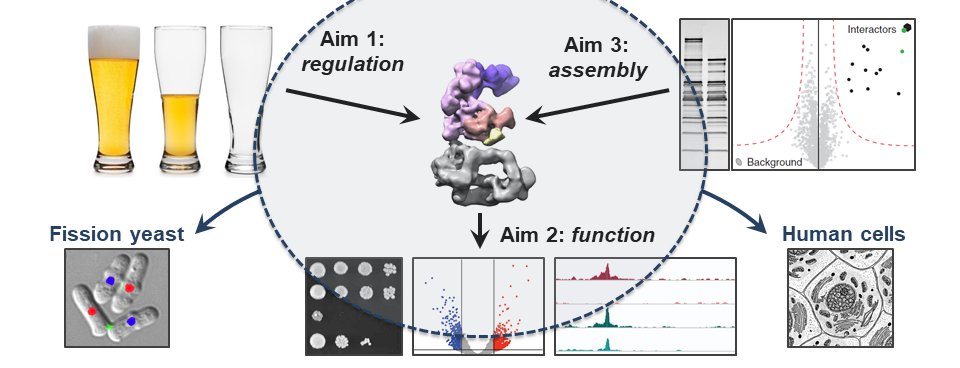
To address these issues, we combine classical molecular, cell biology, genetics, and biochemistry assays with cutting-edge transcriptomic, epigenomic, proteomic, and structural biology approaches, using both the fission yeast Schizosaccharomyces pombe and human cancer cell lines, as experimental systems.
In the presence of nutrients, particularly of a good nitrogen source, S. pombe cells grow and proliferate. In contrast, in starvation conditions, they exit the cell cycle and differentiate into a quiescent state. In this yeast, we decipher the mechanisms by which nutrient-sensing kinases control transcription and chromatin regulators. In parallel, we explore the roles of dedicated chaperones in the coordinated assembly of these factors into active multimeric complexes.
In parallel, we investigate whether similar processes contribute to control the balance between proliferation and quiescence of cancer cells. Indeed, given the high conservation of these factors, what we learn in yeast is likely to be relevant to other, ‘slower-growing’ eukaryotes. This issue is important because some cancer cells could exit quiescence, resume proliferation and cause tumor relapse. Forcing these cells to stay dormant could be an effective way of stopping tumor recurrence following conventional therapies.
Interested in our work? Contact dom [at] helmlinger [dot] com
Follow us on Twitter, LinkedIn, ResearchGate, and ORCID (0000-0003-1501-0423).
En savoir plus
NEWS:
09/2020 – Funding! Our ANR grant ‘COACTIVATOR’ with Nacho Molina and Didier Devys has been accepted!
11/2019 – Paper! Alberto, Damien, and Céline’s work on the assembly of SAGA and NuA4 is now published in Nature Communications. Congrats!
11/2019 – Our perspective on Tra1 evolutionary history, written with the help of Philippe Fort, is published in Biochemical Society Symposia. Congrats!
10/2019 – Welcome to our new PhD student, Pascal Carme, who joins the lab with an CBS2 PhD fellowship!
05/2019 – Funding! Damien obtains a 4th-year PhD fellowship from the Ligue Nationale Contre le Cancer.
02/2019 – Preprint! Alberto’s work on the assembly of Tra1 into SAGA and NuA4 is available as a preprint on biorXiv. Congrats!
02/2019 – Alba Toran-Vilarrubias,from the University of Barcelona, joins the lab for her Master internship – welcome!
01/2019 – Pascal Carme,from the University of Montpellier, joins the lab for his Master internship – welcome!
12/2018 – Funding! ARC « Projets 2018 » to study the role of Hsp90 co-chaperones on colorectal cancer cell resistance to treratment.
11/2018 – Dylane successfully defended her thesis work on the regulation of interferon-stimulated genes by the TRRAP transcription cofactor. Congrats Dr. Detilleux! And best wishes for your postdoctoral work in Strasbourg!
07/2018 – Peggy Raynaud (CR, INSERM) joins the lab – welcome!
02/2018 – Talha Magat, from the University of Montpellier, joins the lab for his 1st year Master internship – welcome!
10/2017 – Paper! Thomas Laboucarié’s work on the regulation of SAGA by the TOR kinase pathways is published in EMBO Reports. Congrats!
09/2017 – Paper! Our review with László Tora on « Sharing the SAGA » is out in Trends in Biochemical Sciences.
05/2017 – Funding! Dylane obtains a 4th-year PhD fellowship from the Ligue Nationale Contre le Cancer.
02/2017 – Diana Bachrouche from the Lebanese University joins the lab for her Master thesis – welcome!
10/2016 – Welcome to our new PhD student, Damien Toullec, who joins the lab with an EpiGenMed PhD fellowship!
06/2016 – Funding! INCa grant awarded together with E. Bertrand and M. Hahne teams @IGMM.
04/2016 – Thomas successfully defended his thesis on the regulation of SAGA by the mTOR signaling pathways in fission yeast. Congrats Dr. Laboucarié, the first PhD graduate of the lab. And best wishes in the vineyards!
01/2016 – Damien Toullec from the University of Rennes joins the lab for his Master internship – welcome!
09/2015 – Paper! Alberto’s work the Hos2 HDAC in Ustilago maydis is now published in PLOS Pathogens.
08/2015 – Funding! Two ANR grants awarded: ANR JCJC « CoCoNut » and PRC « SAGA2 ».
07/2015 – Dom obtains an HDR diploma from the University of Montpellier.
07/2015 – Céline Faux joins the lab as a lab manager on a CNRS permanent position – welcome!
05/2015 – Funding! Thomas obtains a 4th-year PhD fellowship from the Fondation ARC.
03/2015 – Kerstin Wagner from the University of Vienna joins the lab for her Master thesis – welcome!
12/2014 – Dylane Detilleux got a PhD fellowship from the Ligue Nationale Contrer le Cancer and joins the lab!
10/2014 – Bérengère Pradet-Balade (CR1, CNRS) got the HDR diploma from the University of Montpellier and joins the lab – welcome!
12/2013 – Funding! ARC « Projets 2013 »; ATIP grant extended; Marie Curie Career Integration Grants.
11/2013 – Dylane Detilleux from the University of Montpellier joins the lab for her Master internship – welcome!
10/2013 – William Johnsonfrom the University of Manchester joins the lab as part of his Erasmus exchange program – welcome!
07/2013 – Alberto Elias-Villalobos from Sevilla University got a postdoc fellowship from the FRM and joins the lab – welcome!
06/2013 – Funding! ‘Emergences’ grant from the Cancéropôle Grand Sud-Ouest.
10/2012 – Thomas Laboucarié got an EpiGenMed PhD fellowship and joins the lab – welcome!
07/2012 – Yves Romeo got a tenured lecturer position at the University of Toulouse – congrats!
01/2012 – Paper! Dom’s review on Tra1 in buddng and fission yeast is now published in Transcription.
12/2011 – Yves Romeo from McGill University got a postdoc fellowship and joins the lab – welcome!
01/08/2011: Opening Day and first member to join – Gwenda Lledo! Yes we can!
Financements
ANR EPISPORE (Dominique Helmlinger)
ANR SAGA-TBP (Dominique Helmlinger)
ANR COACTIVATOR (Dominique Helmlinger)
80Prime (Dominique Helmlinger (INSB) & Maryse Vanderplanck (INEE))
Publications
2024
- Agar lot-specific inhibition in the plating efficiency of yeast spores and cells. Protacio RU, Davidson MK, Malone EG, Helmlinger D, Smith JR, Gibney PA, Wahls WP. G3 (Bethesda). 2024 Sep 23;14(12):jkae229. Pubmed
- Laboratory horror stories: Poison in the agars. Davidson MK, Protacio RU, Helmlinger D, Wahls WP. bioRxiv [Preprint]. 2024 Jun 6:2024.06.06.597796. Pubmed
2023
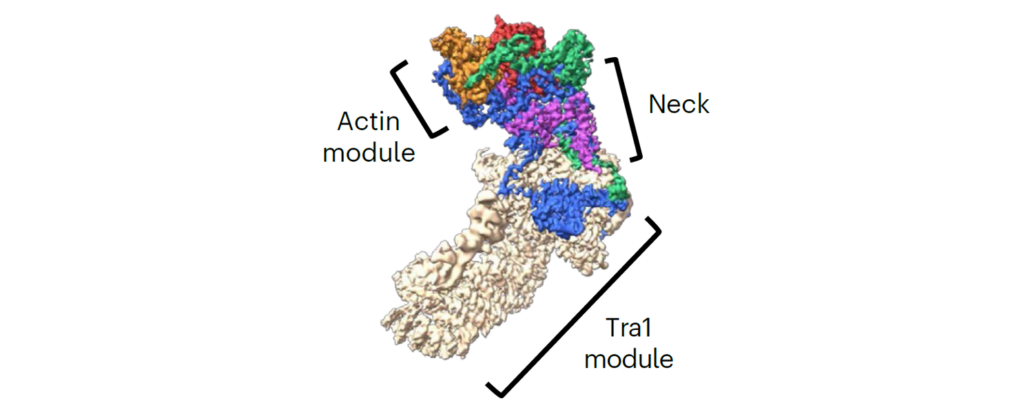
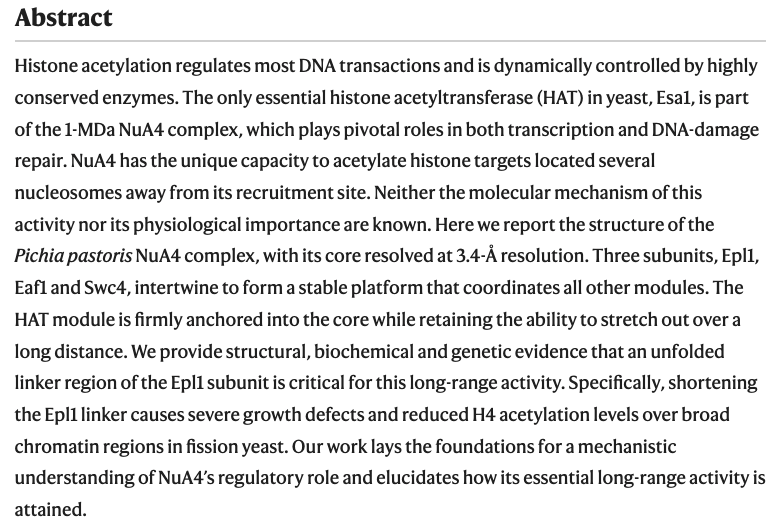
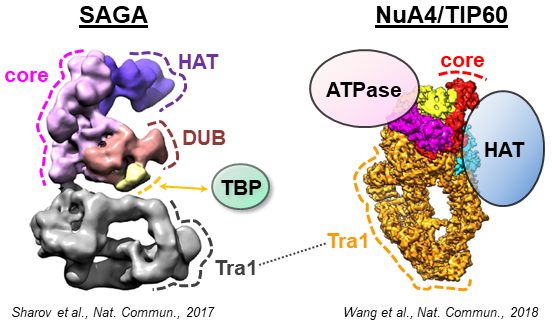
- The structure of the NuA4-Tip60 complex reveals the mechanism and importance of long-range chromatin modification. Fréchard A, Faux C, Hexnerova R, Crucifix C, Papai G, Smirnova E, McKeon C, Ping FLY, Helmlinger* D, Schultz* P, Ben-Shem* A. Nat Struct Mol Biol. 2023 Sep;30(9):1337-1345. Pubmed
- The SAGA histone acetyltransferase module targets SMC5/6 to specific genes. Mahrik L, Stefanovie B, Maresova A, Princova J, Kolesar P, Lelkes E, Faux C, Helmlinger D, Prevorovsky M, Palecek JJ. Epigenetics Chromatin. 2023 Feb 16;16(1):6. Pubmed
2022
- SUPT3H-less SAGA coactivator can assemble and function without significantly perturbing RNA polymerase II transcription in mammalian cells. Fischer V, Hisler V, Scheer E, Lata E, Morlet B, Plassard D, Helmlinger D, Devys D, Tora L, Vincent SD. Nucleic Acids Res. 2022 Aug 12;50(14):7972-7990. Pubmed
- Quantitative analysis of protein-RNA interactions in fission yeast. Elías-Villalobos A, Duncan C, Mata J, Helmlinger D. STAR Protoc. 2022 May 9;3(2):101373. Pubmed
- The TRRAP transcription cofactor represses interferon-stimulated genes in colorectal cancer cells. Detilleux D, Raynaud P, Pradet-Balade B, Helmlinger D. Elife. 2022 Mar 4;11:e69705. Pubmed
2021
- The Hsp90 cochaperone TTT promotes cotranslational maturation of PIKKs prior to complex assembly. Toullec D, Elías-Villalobos A, Faux C, Noly A, Lledo G, Séveno M, Helmlinger D. Cell Rep. 2021 Oct 19;37(3):109867. Pubmed
- The HSP90/R2TP assembly chaperone promotes cell proliferation in the intestinal epithelium. Maurizy C, Abeza C, Lemmers B, Gabola M, Longobardi C, Pinet V, Ferrand M, Paul C, Bremond J, Langa F, Gerbe F, Jay P, Verheggen C, Tinari N, Helmlinger D, Lattanzio R, Bertrand E, Hahne M, Pradet-Balade B. Nat Commun. 12(1):4810. Pubmed
- What do the structures of GCN5-containing complexes teach us about their function? Helmlinger D, Papai G, Devys D, Tora L. Biochim Biophys Acta Gene Regul Mech. 1864(2):194614 Pubmed
2019
- Chaperone-mediated ordered assembly of the SAGA and NuA4 transcription co-activator complexes in yeast. Elías-Villalobos A, Toullec D, Faux C, Séveno M, Helmlinger D. Nat Commun. 10(1):5237 Pubmed
- New insights into the evolutionary conservation of the sole PIKK pseudokinase Tra1/TRRAP. Elías-Villalobos A, Fort P, Helmlinger D. Biochem Soc Trans. 2019 Nov 26. Pubmed
2017
- TORC1 and TORC2 converge to regulate the SAGA co-activator in response to nutrient availability. Laboucarié T, Detilleux D, Rodriguez-Mias RA, Faux C, Romeo Y, Franz-Wachtel M, Krug K, Maček B, Villén J, Petersen J, Helmlinger D. EMBO Rep. pii: e201744942. Pubmed
- Sharing the SAGA. Helmlinger D, Tora L. Trends Biochem Sci. S0968-0004(17)30168-8. Pubmed
2015
- The Hos2 Histone Deacetylase Controls Ustilago maydis Virulence through Direct Regulation of Mating-Type Genes. Elías-Villalobos A, Fernández-Álvarez A, Moreno-Sánchez I, Helmlinger D, Ibeas JI. PLoS Pathog. 11:e1005134. Pubmed
2014
- Drosophila Spag is the homolog of RNA polymerase II-associated protein 3 (RPAP3) and recruits the heat shock proteins 70 and 90 (Hsp70 and Hsp90) during the assembly of cellular machineries. Benbahouche Nel H, Iliopoulos I, Török I, Marhold J, Henri J, Kajava AV, Farkaš R, Kempf T, Schnölzer M, Meyer P, Kiss I, Bertrand E, Mechler BM, Pradet-Balade B. J Biol Chem. 289:6236-47. Pubmed
2012
- HSP90 and the R2TP co-chaperone complex: building multi-protein machineries essential for cell growth and gene expression. Boulon S, Bertrand E, Pradet-Balade B. RNA Biol. 9(2):148-54. Pubmed
- New insights into the SAGA complex from studies of the Tra1 subunit in budding and fission yeast. Helmlinger D. Transcription. 3(1):13-8. Pubmed
2011
- CRM1 controls the composition of nucleoplasmic pre-snoRNA complexes to licence them for nucleolar transport. Pradet-Balade B, Girard C, Boulon S, Paul C, Azzag K, Bordonné R, Bertrand E, Verheggen C. EMBO J. 30(11):2205-18. Pubmed
- Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. Helmlinger D, Marguerat S, Villén J, Swaney DL, Gygi SP, Bähler J, Winston F. EMBO J. 30(14):2843-52. Pubmed
2010
- HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, Lamond AI, Bertrand E. Mol Cell. 39(6):912-924. Pubmed
2008
- The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Helmlinger D, Marguerat S, Villén J, Gygi SP, Bähler J, Winston F. Genes Dev. 22(22):3184-95. Pubmed
Regulation of gene expression
 Dominique HELMLINGER
Dominique HELMLINGER
Group leader (Research Director CNRS)
Team Members
(Chercheur DR2) (+33) 04 34 35 95 48 |
|
(Émérite) +33 (0)4 34 35 95 00 |
|
(CRCN) +33 (0)4 34 35 95 49 |
|
(Chercheur DR2) +33 (0)4 34 35 95 51 |
|
(IE-Recherche) +33 (0)4 34 35 95 51 |
|
(Doctorant) +33 (0)4 34 35 95 49 |
|
(Doctorant) +33 (0)4 34 35 95 51 |
|
(AI-Recherche) +33 (0)4 34 35 95 49 |
|
(Doctorant) +33 (0)4 34 35 95 49 |

Contact us
Replace the name and address above with that of the member to contact
firstname.name@crbm.cnrs.fr