Our Research
The genetic material is the substrate of transactions such as transcription and replication, which engender the risk of creating lesions on it. Moreover, the DNA is exposed to both external and internal sources of damage. In all of these instances, the cell sets into action surveillance and repair mechanisms to preserve the integrity of the genome. All these events take place within the nucleus of the cell, and therefore much attention and research efforts have been devoted to deeply understand these nuclear mechanisms. Yet, the nucleus is embedded in the cytoplasm, and recent investigations start to shed light onto the impact that processes up-to-here considered to be exclusively cytoplasmic may have on the organization, maintenance and dynamics of the genome. The aim of our team is to enlarge our knowledge of how nutritional setups, environmental cues and metabolic alterations affect or even control genome integrity maintenance.
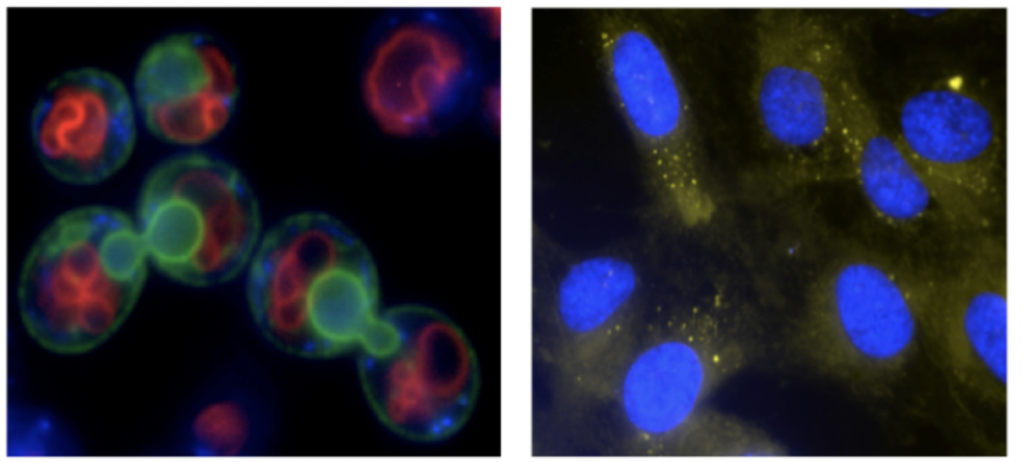
In particular, we focus on the participation of lipids. Lipids are a vast and versatile family of biomolecules known for their essential roles as membrane constituents, source of energy and signaling abilities. Yet, whether these functions (or others) are important for nuclear homeostasis, in general, and to genome stability surveillance and maintenance, in particular, remains almost unexplored. We take advantage of the yeast Sacharomyces cerevisiae and of cultured human cells as model systems. We utilize a combination of genetics, molecular biology, biochemistry and microscopy to assess these questions.
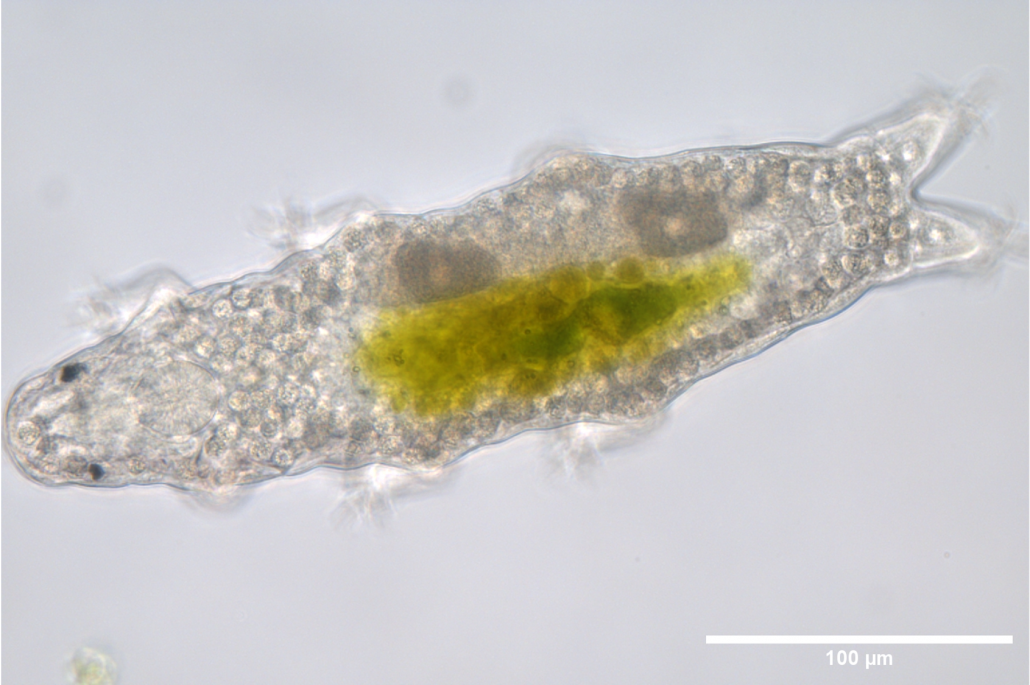
Additionally, to further expand our insights into the detection of DNA damages and the regulation of this process, we have recently incorporated to our laboratory the tardigrade experimental model Hypsibius exemplaris. Tardigrades are extremophile organisms known to endure truly harsh conditions such as extreme temperatures, dehydration, and ionizing radiation (IR). In particular, H. exemplaris is strongly resistant to IR in its adult (and hydrated) stage, thus displaying a remarkable DNA damage resistance compared to most organisms. We use H. exemplaris to improve our understanding on the cellular and molecular basis of this extraordinary DNA damage tolerance. H. exemplaris is an excellent model, as it possesses a sequenced genome, can be studied at the molecular level thanks to well-established protocols, has a short life cycle, and is transparent, helping us in microscopy-based approaches.
Funding
Publications
2025
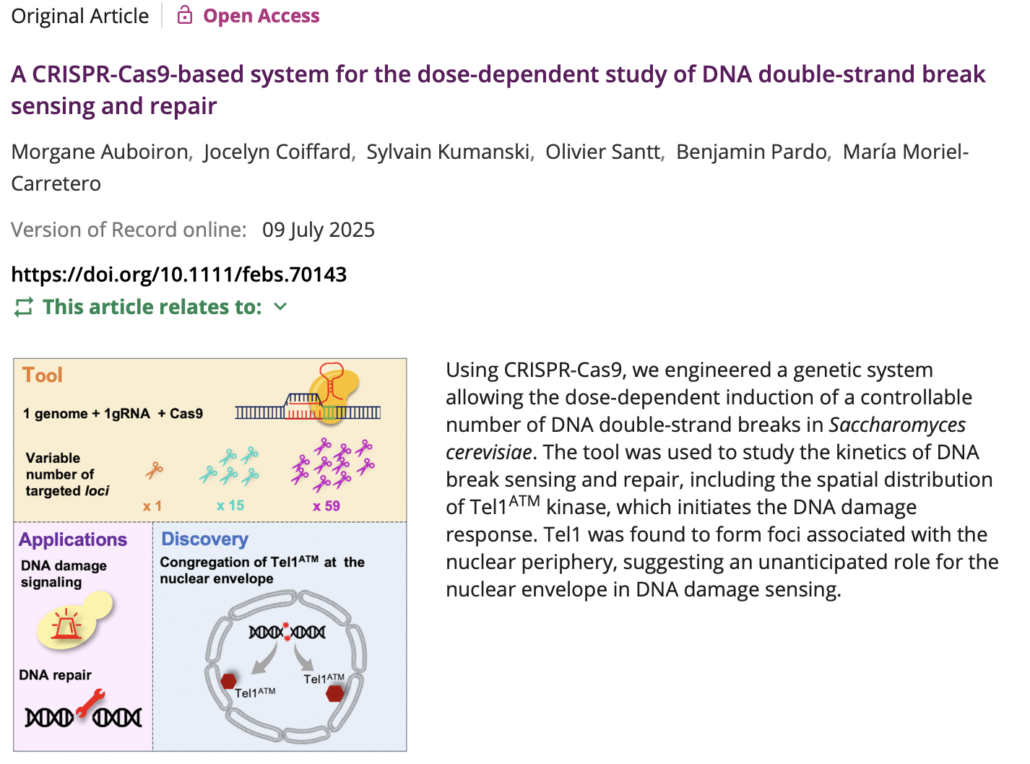
- A CRISPR-Cas9-based system for the dose-dependent study of DNA double-strand break sensing and repair. Auboiron M, Coiffard J, Kumanski S, Santt O, Pardo B, Moriel-Carretero M. FEBS J. 2025 Jul 9. Epub ahead of print. PMID: 40631397 Pubmed
- Neutral lipids restrict the mobility of broken DNA molecules during comet assays. Soulet C, Josa-Castro J, Moriel-Carretero M. Biol Cell. 2025 Jan;117(1):e2400141. Pubmed
2024
- Behind the stoNE wall: a fervent activity for nuclear lipids. Samardak K, Bâcle J, Moriel-Carretero M. Biochimie. 2024 Aug 5:S0300-9084(24)00179-2. Pubmed
- Storage cell proliferation during somatic growth establishes that tardigrades are not eutelic organisms. Quiroga-Artigas G, Moriel-Carretero M. Biol Open. 2024 Feb 15;13(2):bio060299.. Pubmed
2023
- An Expansion of the Endoplasmic Reticulum that Halts Autophagy is Permissive to Genome Instability. Lara-Barba E, Torán-Vilarrubias A, Moriel-Carretero M. Contact. 2023;6. Pubmed
- Nuclear envelope-remodeling events as models to assess the potential role of membranes on genome stability. Bâcle J, Groizard L, Kumanski S, Moriel-Carretero M. FEBS Lett. 2023 Jun 20. Pubmed
- A sterol-PI(4)P exchanger modulates the Tel1/ATM axis of the DNA damage response. Ovejero S, Kumanski S, Soulet C, Azarli J, Pardo B, Santt O, Constantinou A, Pasero P, Moriel-Carretero M. EMBO J. 2023 Jun 12:e112684. Pubmed
- Auxin alone provokes retention of ASH1 mRNA in Saccharomyces cerevisiae mother cells. Domeni Zali G, Moriel-Carretero M. MicroPubl Biol. 2023 Feb 22;2023:10.17912/micropub.biology.000752.Pubmed
2022
- Nuclear ingression of cytoplasmic bodies accompanies a boost in autophagy. Garcia M, Kumanski S, Elías-Villalobos A, Cazevieille C, Soulet C, Moriel-Carretero M. Life Sci Alliance. 2022 May 13;5(9):e202101160. Pubmed
- Coordination between phospholipid pools and DNA damage sensing. Ovejero S, Soulet C, Kumanski S, Moriel-Carretero M. Biol Cell. 2022 May 7. Pubmed
- Nuclear Lipid Droplet Birth during Replicative Stress. Kumanski S, Forey R, Cazevieille C, Moriel-Carretero M. Cells. 2022 11, 1390. Pubmed
2021
- The Many Faces of Lipids in Genome Stability (and How to Unmask Them). Moriel-Carretero M. Int J Mol Sci. 2021 Nov 29;22(23):12930. Pubmed
- Lack of evidence for condensin or cohesin sequestration on lipid droplets with packing defects. Mura A, Moriel-Carretero M. MicroPubl Biol. 2021 Nov 4;2021. Pubmed
- Microscopy analysis of the smallest subunit of the RPA complex, Rfa3p, prompts consideration of how RPA subunits gather at single-stranded DNA sites. Ramonatxo A, Moriel-Carretero M. MicroPubl Biol. 2021 Oct 27:2021:10.17912/micropub.biology.000493. Pubmed
- Oxidative agents elicit endoplasmic reticulum morphological changes suggestive of alterations in lipid metabolism. Torán-Vilarrubias A, Moriel-Carretero M. MicroPubl Biol. 2021 Sep 20 (eCollection 2021) Pubmed
- Daughter cell-targeted mRNAs can achieve segregation without the universal Endoplasmic Reticulum docker She2p. Samardak K, Moriel-Carretero M. MicroPubl Biol. 2021 Sep 13 (eCollection 2021) Pubmed
- The Alkylating Agent Methyl Methanesulfonate Triggers Lipid Alterations at the Inner Nuclear Membrane That Are Independent from Its DNA-Damaging Ability. Ovejero S, Soulet C, Moriel-Carretero M. Int J Mol Sci. 22(14):7461. Pubmed
- Lipid Droplets Are a Physiological Nucleoporin Reservoir. Kumanski S, Viart BT, Kossida S, Moriel-Carretero M. Cells. 10(2):472. Pubmed
2020
- Homologous Recombination and Mus81 Promote Replication Completion in Response to Replication Fork Blockage. Benjamin Pardo, María Moriel-Carretero, Thibaud Vicat, Andrés Aguilera , Philippe Pasero. EMBO Rep. 17;e49367 Pubmed
- The Hypothetical Role of Phosphatidic Acid in Subverting ER Membranes During SARS-CoV Infection. María Moriel-Carretero. Traffic . 2020 May 18. doi: 10.1111/tra.12738 Pubmed
2019
Cytoplasmic control of genome stability
Team Members
(Chercheur) +33 (0)4 34 35 95 64 |
|
(Doctorant) +33 (0)4 34 35 95 64 |
|
(Chercheur) +33 (0)4 34 35 96 83 |
|
(Doctorant) +33 (0)4 34 35 96 83 |
|
(IR-Recherche) +33 (0)4 34 35 96 83 |
|
(IE-Recherche) +33 (0)4 34 35 95 64 |
|
(Stagiaire) +33 (0)4 34 35 9 |
team

team
Contact us
Replace the name and address below with that of the member to contact
firstname.name@crbm.cnrs.fr
team