Our Research
During cell division, chromosomes are faithfully duplicated and segregated, so that one copy of each chromosome can be inherited by the two daughter cells, which must carry identical genetic information. The fidelity of chromosome transmission has important medical implications. For example, aberrant chromosome numbers in eggs or sperms generate embryos that undergo spontaneous miscarriage or that have severe pathologies, such as the Down syndrome. Inaccurate genome partitioning during any of the millions of cell divisions taking place in our body can lead to tumour development.
Our laboratory studies the surveillance mechanisms that act during mitosis to ensure proper genome partitioning, using the budding yeast Saccharomyces cerevisiae as a model organism. Particularly, we focus on two aspects of mitosis that are crucial for preventing aneuploidy formation:
- how faithful chromosome segregation is overseen by the spindle assembly checkpoint (SAC) that monitors the correct attachment of chromosomes to the mitotic spindle;
- how mitotic exit and cytokinesis are regulated to preserve genome stability.
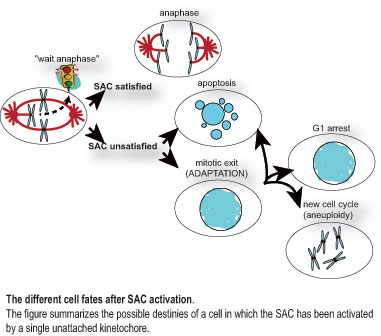
Mechanisms sustaining the Spindle Assembly Checkpoint (SAC)
The SAC prevents anaphase when errors occur during the bi-orientation of chromosomes on the mitotic spindle. By delaying sister chromatid separation and mitotic exit, the SAC gives to the cell the time necessary to establish the appropriate connections between kinetochores and microtubules, thus ensuring balanced chromosome partitioning. Once all chromosomes are bi-oriented, the SAC is turned off, allowing anaphase to take place.
Despite its importance, SAC-induced cell cycle arrest is transient. Moreover, if the SAC remains activated for a prolonged time, cells will escape mitotic arrest and adapt, through a process known as “mitotic slippage”. Mitotic slippage is thought to be a major cause of resistance of cancer cells to chemotherapeutic compounds that interfere with microtubule dynamics.
Despite the profound implications of mitotic slippage for human health, the factors influencing its speed and efficiency are largely unknown. In our laboratory we use genetic screens to identify novel genes involved in this process and to characterize the molecular mechanism(s) through which they control slippage.
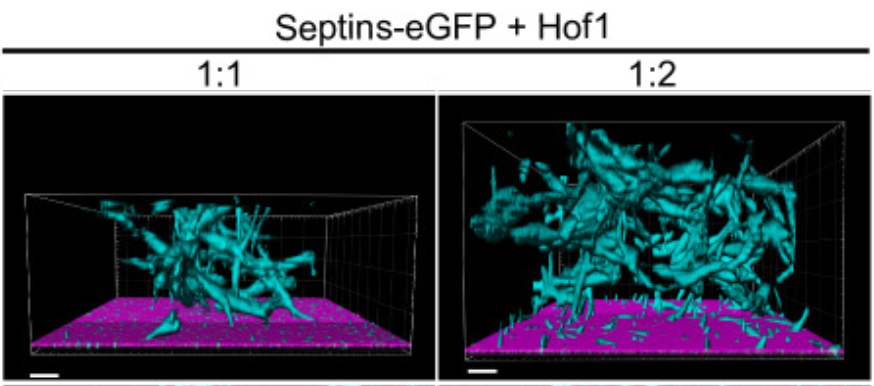
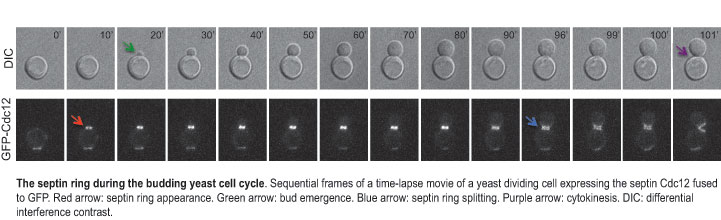
Regulation of septin dynamics during cytokinesis
Growing evidence indicates that polyploid cells are genetically unstable and frequently undergo chromosome missegregation and aneuploidy, eventually leading to cancer development. Cytokinesis failure is a common mechanism leading to polyploidy.
In many animal cells and fungi, cytokinesis requires septins and a contractile actomyosin ring. In yeast, septins form a ring at the division site throughout most the cell cycle. However, the septin ring dynamics changes during the cell cycle. Particularly, just prior to cytokinesis, a spectacular remodelling of the septin ring occurs. This involves its splitting in two separate rings that sandwich the constricting actomyosin ring. Our laboratory is trying to identify and characterize, through genetic and proteomic approaches, the molecular determinants that mediate the septin ring dynamic transitions during the cell cycle.
Read more
Funding
FRM (Simonetta Piatti)
ANR SEPTORG (Simonetta Piatti)
Publications
2025
- Septin assemblies promote the lipid organization of membranes. El Alaoui F, Al-Akiki I, Ibanes S, Lyonnais S, Sanchez-Fuentes D, Desgarceaux R, Cazevieille C, Blanchard MP, Parmeggiani A, Carretero-Genevrier A, Piatti S, Picas L. Structure. 2025 Mar 6;33(3):451-464.e5. Pubmed
2024
- Septin Organization and Dynamics for Budding Yeast Cytokinesis. Varela Salgado M, Piatti S. J Fungi (Basel). 2024 Sep 9;10(9):642. Pubmed
- Phosphorylation of the F-BAR protein Hof1 drives septin ring splitting in budding yeast. Salgado MV, Adriaans IE, Touati SA, Ibanes S, Lai-Kee-Him J, Ancelin A, Cipelletti L, Picas L and Piatti S.. Nat Commun 15, 3383 Open Access
2022
- The Syp1/FCHo2 protein induces septin filament bundling through its intrinsically disordered domain. Ibanes S, El-Alaoui F, Lai-Kee-Him J, Cazevieille C, Hoh F, Lyonnais S, Bron P, Cipelletti L, Picas L, Piatti S. Cell Rep. 41(10):111765. Pubmed
2021
- Downregulation of the Tem1 GTPase by Amn1 after cytokinesis involves both nuclear import and SCF-mediated degradation. Devault A, Piatti S. J Cell Sci. jcs.258972 Pubmed
2020
- Silencing the spindle assembly checkpoint: Let’s play Polo! Benzi G, Piatti S. J Cell Biol. 219(12):e202010053 Pubmed
- Killing two birds with one stone: how budding yeast Mps1 controls chromosome segregation and spindle assembly checkpoint through phosphorylation of a single kinetochore protein. Giorgia Benzi, Simonetta Piatti. Curr Genet. 66(6):1037-1044 Pubmed
- Cytokinesis: An Anillin-RhoGEF Module Sets the Stage for Septin Double Ring Assembly. Piatti S. Curr Biol. 30(8):R347-R349 Pubmed
- A common molecular mechanism underlies the role of Mps1 in chromosome biorientation and the spindle assembly checkpoint. Benzi G, Camasses A, Atsunori Y, Katou Y, Shirahige K, Piatti S. EMBO Rep. e50257. Pubmed
- The Phosphatase PP1 Promotes Mitotic Slippage through Mad3 Dephosphorylation. Ruggiero A, Katou Y, Shirahige K, Séveno M, Piatti S. Curr Biol. 30(2):335-343.e5. Pubmed
2019
- Septin clearance from the division site triggers cytokinesis in budding yeast. Tamborrini D, Piatti S. Microb Cell. 6(6):295-298. Pubmed
2018
- Recruitment of the mitotic exit network to yeast centrosomes couples septin displacement to actomyosin constriction. Tamborrini D, Juanes MA, Ibanes S, Rancati G, Piatti S. Nat Commun. 9(1):4308. Pubmed
2017
- Asymmetric Localization of Components and Regulators of the Mitotic Exit Network at Spindle Pole Bodies. Scarfone I, Piatti S. Methods Mol Biol. 1505:183-193. Pubmed
2016
- Control of Formin Distribution and Actin Cable Assembly by the E3 Ubiquitin Ligases Dma1 and Dma2. Juanes MA, Piatti S. Genetics. 204:205-20. Pubmed
- The final cut: cell polarity meets cytokinesis at the bud neck in S. cerevisiae. Juanes MA, Piatti S. Cell Mol Life Sci. 73:3115-36. Pubmed
2015
- Asymmetry of the budding yeast Tem1 GTPase at spindle poles is required for spindle positioning but not for mitotic exit. Scarfone I, Venturetti M, Hotz M, Lengefeld J, Barral Y, Piatti S. PLoS Genet. 11:e1004938. Pubmed
- Coupling spindle position with mitotic exit in budding yeast: The multifaceted role of the small GTPase Tem1. Scarfone I, Piatti S. Small GTPases. 6:196-201. Pubmed
- Rho1- and Pkc1-dependent phosphorylation of the F-BAR protein Syp1 contributes to septin ring assembly. Merlini L, Bolognesi A, Juanes MA, Vandermoere F, Courtellemont T, Pascolutti R, Séveno M, Barral Y, Piatti S. Mol Biol Cell. 26:3245-62. Pubmed
2013
- Budding yeast greatwall and endosulfines control activity and spatial regulation of PP2A(Cdc55) for timely mitotic progression. Juanes MA, Khoueiry R, Kupka T, Castro A, Mudrak I, Ogris E, Lorca T, Piatti S. PLoS Genet. 9(7):e1003575. Pubmed
- Yeast haspin kinase regulates polarity cues necessary for mitotic spindle positioning and is required to tolerate mitotic arrest. Panigada D, Grianti P, Nespoli A, Rotondo G, Castro DG, Quadri R, Piatti S, Plevani P, Muzi-Falconi M. Dev Cell. 26(5):483-95. Pubmed
2012
- Budding yeast dma proteins control septin dynamics and the spindle position checkpoint by promoting the recruitment of the Elm1 kinase to the bud neck. Merlini L, Fraschini R, Boettcher B, Barral Y, Lucchini G, Piatti S. PLoS Genet. 8(4):e1002670. Pubmed
- Role of the Mad2 dimerization interface in the spindle assembly checkpoint independent of kinetochores. Mariani L, Chiroli E, Nezi L, Muller H, Piatti S, Musacchio A, Ciliberto A. Curr Biol. 22(20):1900-8. Pubmed
2011
- Analysis of the rpn11-m1 proteasomal mutant reveals connection between cell cycle and mitochondrial biogenesis. Esposito M, Piatti S, Hofmann L, Frontali L, Delahodde A, Rinaldi T. FEMS Yeast Res. 11(1):60-71. Pubmed
- The mother-bud neck as a signaling platform for the coordination between spindle position and cytokinesis in budding yeast. Merlini L, Piatti S. Biol Chem. 392(8-9):805-12. Pubmed
2010
- The RSC chromatin-remodeling complex influences mitotic exit and adaptation to the spindle assembly checkpoint by controlling the Cdc14 phosphatase. Rossio V, Galati E, Ferrari M, Pellicioli A, Sutani T, Shirahige K, Lucchini G, Piatti S. J Cell Biol. 191(5):981-97. Pubmed
- Adapt or die: how eukaryotic cells respond to prolonged activation of the spindle assembly checkpoint. Rossio V, Galati E, Piatti S. Biochem Soc Trans. 38(6):1645-9. Pubmed
2009
- Cdc14 inhibition by the spindle assembly checkpoint prevents unscheduled centrosome separation in budding yeast. Chiroli E, Rancati G, Catusi I, Lucchini G, Piatti S. Mol Biol Cell. 20(10):2626-37. Pubmed
2008
- The spindle position checkpoint: how to deal with spindle misalignment during asymmetric cell division in budding yeast. Fraschini R, Venturetti M, Chiroli E, Piatti S. Biochem Soc Trans. 36(Pt 3):416-20. Pubmed
Mitotic regulation of chromosome partitioning and cell division
Simonetta PIATTI
Group Leader (Research Director CNRS)
Team Members
(IE-Recherche) (+33) 04 34 35 95 45 |
|
(Chercheur DR1) +33 (0)4 34 35 95 46 |
|
(IE-Recherche) +33 (0)4 34 35 95 48 |
|
(Doctorant) +33 (0)4 34 35 95 45 |
|
(Doctorant) +33 (0)4 34 35 95 45 |
|
(Stagiaire) +33 (0)4 34 35 9 |

Contact us
Replace the name and address below with that of the member to contact
firstname.name@crbm.cnrs.fr